You are browsing environment: FUNGIDB
CAZyme Information: SHO77213.1
You are here: Home > Sequence: SHO77213.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
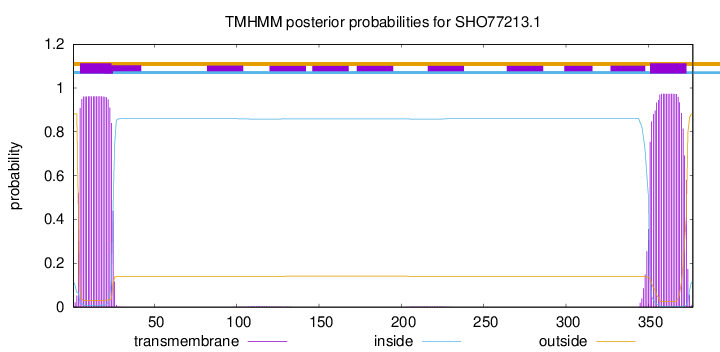
TMHMM annotations
Basic Information help
| Species | Malassezia sympodialis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Malasseziomycetes; ; Malasseziaceae; Malassezia; Malassezia sympodialis | |||||||||||
| CAZyme ID | SHO77213.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | GH16 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1M8A424] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 61 | 315 | 6e-64 | 0.9868995633187773 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185690 | GH16_fungal_Lam16A_glucanase | 8.18e-83 | 29 | 335 | 9 | 292 | fungal 1,3(4)-beta-D-glucanases, similar to Phanerochaete chrysosporium laminarinase 16A. Group of fungal 1,3(4)-beta-D-glucanases, similar to Phanerochaete chrysosporium laminarinase 16A. Lam16A belongs to the 'nonspecific' 1,3(4)-beta-glucanase subfamily, although beta-1,6 branching and beta-1,4 bonds specifically define where Lam16A hydrolyzes its substrates, like curdlan (beta-1,3-glucan), lichenin (beta-1,3-1,4-mixed linkage glucan), and laminarin (beta-1,6-branched-1,3-glucan). |
| 185693 | GH16_laminarinase_like | 6.38e-07 | 116 | 159 | 88 | 132 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| 185691 | GH16_Strep_laminarinase_like | 1.63e-04 | 123 | 157 | 108 | 148 | Streptomyces laminarinase-like, member of glycosyl hydrolase family 16. Proteins similar to Streptomyces sioyaensis beta-1,3-glucanase (laminarinase) present in Actinomycetales as well as Peziomycotina. Laminarinases belong to glycosyl hydrolase family 16 and hydrolyze the glycosidic bond of the 1,3-beta-linked glucan, a major component of fungal and plant cell walls and the structural and storage polysaccharides (laminarin) of marine macro-algae. Members of the GH16 family have a conserved jelly roll fold with an active site channel. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.57e-291 | 1 | 377 | 1 | 377 | |
| 8.25e-128 | 6 | 354 | 1 | 355 | |
| 8.25e-128 | 6 | 354 | 1 | 355 | |
| 4.18e-100 | 18 | 343 | 24 | 372 | |
| 6.10e-97 | 7 | 337 | 12 | 365 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.84e-55 | 30 | 335 | 11 | 297 | Chain A, PUTATIVE LAMINARINASE [Phanerodontia chrysosporium],2W39_A Chain A, Putative Laminarinase [Phanerodontia chrysosporium],2W52_A Chain A, PUTATIVE LAMINARINASE [Phanerodontia chrysosporium] |
|
| 4.32e-54 | 30 | 335 | 11 | 297 | Chain A, PUTATIVE LAMINARINASE [Phanerodontia chrysosporium],2WNE_A Chain A, PUTATIVE LAMINARINASE [Phanerodontia chrysosporium] |
|
| 1.49e-40 | 21 | 335 | 2 | 297 | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase [Paecilomyces sp. 'thermophila'],5JVV_B Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase [Paecilomyces sp. 'thermophila'] |
|
| 1.52e-39 | 30 | 336 | 11 | 296 | The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'],3WDU_B The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'],3WDU_C The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'],3WDU_D The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'] |
|
| 1.56e-39 | 30 | 336 | 12 | 297 | The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDT_B The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDT_C The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDT_D The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDV_A The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDV_B The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDV_C The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDV_D The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.67e-44 | 21 | 335 | 128 | 417 | Probable glycosidase C21B10.07 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC21B10.07 PE=3 SV=1 |
|
| 9.96e-39 | 33 | 335 | 31 | 314 | Endo-1,3(4)-beta-glucanase ARB_04519 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_04519 PE=3 SV=1 |
|
| 4.07e-38 | 20 | 348 | 25 | 333 | Probable endo-1,3(4)-beta-glucanase An02g00850 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=An02g00850 PE=3 SV=1 |
|
| 5.39e-38 | 20 | 335 | 27 | 323 | Endo-1,3(4)-beta-glucanase xgeA OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=3 SV=1 |
|
| 1.67e-37 | 21 | 335 | 31 | 326 | Probable endo-1,3(4)-beta-glucanase AFUB_029980 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_029980 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002903 | 0.997056 | CS pos: 19-20. Pr: 0.9665 |