You are browsing environment: FUNGIDB
CAZyme Information: SDRG_16642-t26_1-p1
You are here: Home > Sequence: SDRG_16642-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia diclina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia diclina | |||||||||||
| CAZyme ID | SDRG_16642-t26_1-p1 | |||||||||||
| CAZy Family | GT48 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.53:1 | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA15 | 22 | 205 | 2.5e-54 | 0.9948186528497409 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397269 | LPMO_10 | 1.08e-11 | 99 | 205 | 89 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| 197593 | fCBD | 3.57e-08 | 306 | 339 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 4.33e-08 | 307 | 335 | 1 | 29 | Fungal cellulose binding domain. |
| 282710 | PT | 1.14e-06 | 258 | 292 | 2 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| 282710 | PT | 2.05e-06 | 250 | 282 | 2 | 34 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.91e-212 | 1 | 345 | 1 | 345 | |
| 8.62e-182 | 25 | 345 | 1 | 309 | |
| 4.52e-156 | 19 | 342 | 18 | 318 | |
| 3.69e-135 | 9 | 345 | 7 | 305 | |
| 1.84e-134 | 9 | 345 | 7 | 301 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.20e-22 | 22 | 208 | 1 | 193 | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State [Thermobia domestica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.53e-07 | 308 | 340 | 31 | 63 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=CEL6A PE=1 SV=1 |
|
| 7.38e-07 | 308 | 340 | 32 | 64 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel6A PE=1 SV=1 |
|
| 1.62e-06 | 308 | 340 | 23 | 55 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
|
| 1.62e-06 | 308 | 340 | 23 | 55 | Mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=manF PE=1 SV=2 |
|
| 2.16e-06 | 301 | 343 | 18 | 60 | Exoglucanase 3 OS=Agaricus bisporus OX=5341 GN=cel3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
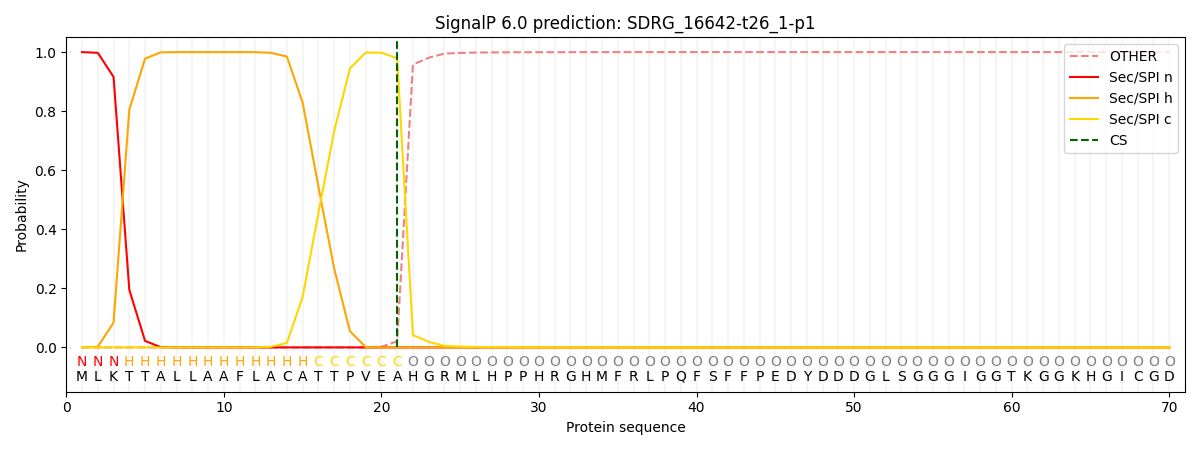
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000286 | 0.999694 | CS pos: 21-22. Pr: 0.9792 |
