You are browsing environment: FUNGIDB
CAZyme Information: SDRG_16535-t26_1-p1
You are here: Home > Sequence: SDRG_16535-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia diclina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia diclina | |||||||||||
| CAZyme ID | SDRG_16535-t26_1-p1 | |||||||||||
| CAZy Family | GT48 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 6 | 219 | 3.2e-71 | 0.8326530612244898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395595 | CBM_1 | 6.72e-07 | 277 | 304 | 2 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 1.79e-06 | 277 | 308 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 273167 | rad23 | 4.35e-04 | 181 | 228 | 77 | 120 | UV excision repair protein Rad23. All proteins in this family for which functions are known are components of a multiprotein complex used for targeting nucleotide excision repair to specific parts of the genome. In humans, Rad23 complexes with the XPC protein. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
| 235540 | PRK05641 | 0.002 | 180 | 233 | 36 | 90 | putative acetyl-CoA carboxylase biotin carboxyl carrier protein subunit; Validated |
| 197593 | fCBD | 0.006 | 232 | 264 | 1 | 33 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.50e-225 | 1 | 313 | 1 | 313 | |
| 3.11e-158 | 4 | 313 | 5 | 321 | |
| 2.02e-87 | 4 | 270 | 8 | 266 | |
| 1.14e-81 | 12 | 264 | 15 | 272 | |
| 3.91e-79 | 1 | 264 | 5 | 272 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.82e-14 | 18 | 184 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
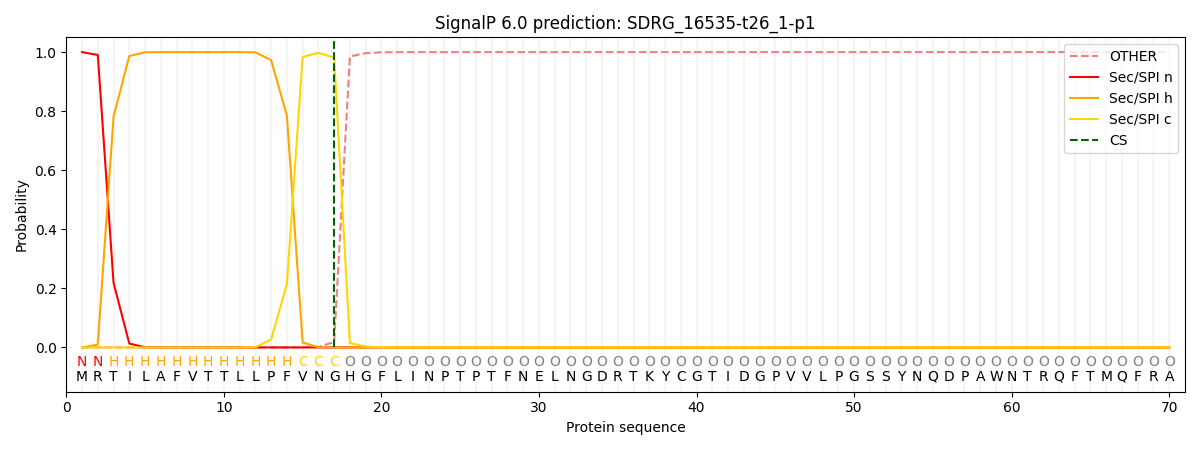
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000282 | 0.999675 | CS pos: 17-18. Pr: 0.9815 |
