You are browsing environment: FUNGIDB
CAZyme Information: SDRG_16162-t26_1-p1
You are here: Home > Sequence: SDRG_16162-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia diclina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia diclina | |||||||||||
| CAZyme ID | SDRG_16162-t26_1-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.53:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA15 | 20 | 204 | 2.1e-56 | 0.9948186528497409 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397269 | LPMO_10 | 3.71e-12 | 20 | 204 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| 395595 | CBM_1 | 8.32e-11 | 306 | 333 | 2 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 9.84e-10 | 304 | 337 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 237864 | PRK14950 | 8.47e-06 | 213 | 279 | 361 | 428 | DNA polymerase III subunits gamma and tau; Provisional |
| 411490 | pneumo_PspA | 1.59e-05 | 214 | 278 | 377 | 439 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.48e-202 | 1 | 342 | 1 | 342 | |
| 1.22e-187 | 1 | 342 | 1 | 337 | |
| 6.35e-168 | 1 | 342 | 1 | 323 | |
| 1.46e-152 | 1 | 340 | 1 | 320 | |
| 1.46e-151 | 1 | 342 | 1 | 309 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.33e-24 | 20 | 207 | 1 | 193 | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State [Thermobia domestica] |
|
| 3.50e-06 | 308 | 337 | 556 | 585 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 3.89e-06 | 308 | 337 | 778 | 807 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.82e-09 | 305 | 338 | 30 | 63 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=CEL6A PE=1 SV=1 |
|
| 4.14e-08 | 306 | 338 | 32 | 64 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel6A PE=1 SV=1 |
|
| 5.11e-08 | 305 | 338 | 24 | 57 | Exoglucanase 3 OS=Agaricus bisporus OX=5341 GN=cel3 PE=1 SV=1 |
|
| 1.83e-07 | 298 | 337 | 498 | 537 | Exoglucanase 1 OS=Penicillium janthinellum OX=5079 GN=cbh1 PE=2 SV=1 |
|
| 2.54e-07 | 305 | 338 | 21 | 54 | Endo-1,4-beta-xylanase 6 OS=Magnaporthe grisea OX=148305 GN=XYL6 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
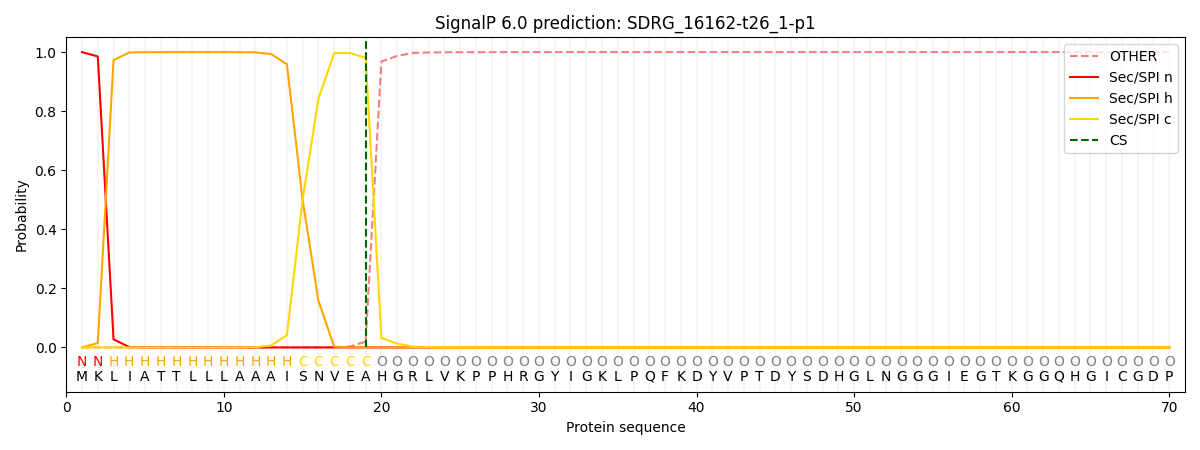
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000220 | 0.999752 | CS pos: 19-20. Pr: 0.9803 |
