You are browsing environment: FUNGIDB
CAZyme Information: SDRG_15701-t26_1-p1
You are here: Home > Sequence: SDRG_15701-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia diclina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia diclina | |||||||||||
| CAZyme ID | SDRG_15701-t26_1-p1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.2:130 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH14 | 24 | 435 | 5.4e-123 | 0.970873786407767 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178400 | PLN02803 | 2.40e-122 | 20 | 450 | 86 | 510 | beta-amylase |
| 215431 | PLN02801 | 6.27e-114 | 20 | 434 | 16 | 428 | beta-amylase |
| 215099 | PLN00197 | 1.06e-112 | 20 | 448 | 106 | 534 | beta-amylase; Provisional |
| 366599 | Glyco_hydro_14 | 4.20e-98 | 24 | 433 | 2 | 388 | Glycosyl hydrolase family 14. This family are beta amylases. |
| 178493 | PLN02905 | 3.53e-95 | 20 | 447 | 265 | 692 | beta-amylase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 450 | 1 | 450 | |
| 0.0 | 1 | 450 | 1 | 450 | |
| 5.53e-222 | 3 | 448 | 4 | 444 | |
| 8.63e-219 | 20 | 448 | 21 | 444 | |
| 4.95e-211 | 8 | 450 | 18 | 456 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.73e-90 | 20 | 406 | 12 | 384 | Chain A, beta-amylase [Glycine max] |
|
| 1.34e-89 | 20 | 406 | 12 | 384 | Beta-AmylaseBETA-Cyclodextrin Complex [Glycine max],1Q6C_A Crystal Structure of Soybean Beta-Amylase Complexed with Maltose [Glycine max],1WDP_A The role of an inner loop in the catalytic mechanism of soybean beta-amylase [Glycine max] |
|
| 1.88e-89 | 20 | 406 | 12 | 384 | Chain A, Beta-amylase [Glycine max],1UKO_B Chain B, Beta-amylase [Glycine max],1UKO_C Chain C, Beta-amylase [Glycine max],1UKO_D Chain D, Beta-amylase [Glycine max] |
|
| 1.88e-89 | 20 | 406 | 12 | 384 | Chain A, Beta-amylase [Glycine max],1UKP_B Chain B, Beta-amylase [Glycine max],1UKP_C Chain C, Beta-amylase [Glycine max],1UKP_D Chain D, Beta-amylase [Glycine max] |
|
| 2.16e-89 | 20 | 406 | 6 | 378 | Chain A, PROTEIN (BETA-AMYLASE) [Hordeum vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.08e-98 | 18 | 450 | 84 | 510 | Beta-amylase 3, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=BAM3 PE=1 SV=3 |
|
| 1.69e-96 | 20 | 448 | 93 | 518 | Beta-amylase 2, chloroplastic OS=Oryza sativa subsp. japonica OX=39947 GN=BAMY2 PE=1 SV=1 |
|
| 2.88e-95 | 20 | 444 | 73 | 495 | Beta-amylase 1, chloroplastic OS=Oryza sativa subsp. japonica OX=39947 GN=BAMY1 PE=1 SV=1 |
|
| 1.11e-91 | 20 | 448 | 106 | 536 | Beta-amylase 1, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=BAM1 PE=1 SV=1 |
|
| 1.44e-88 | 20 | 406 | 10 | 382 | Beta-amylase OS=Hordeum vulgare OX=4513 GN=BMY1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
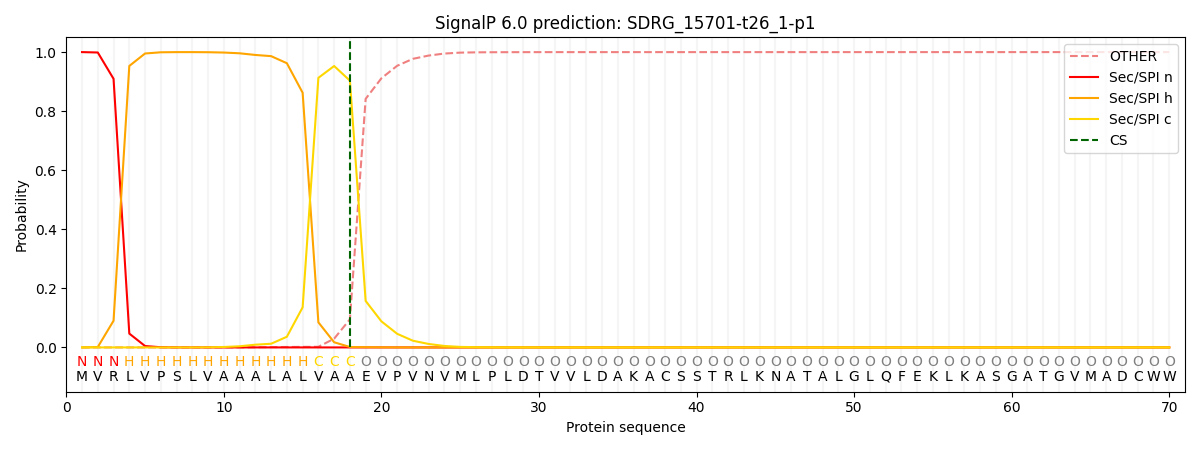
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000527 | 0.999444 | CS pos: 18-19. Pr: 0.9034 |
