You are browsing environment: FUNGIDB
CAZyme Information: SDRG_14951-t26_1-p1
You are here: Home > Sequence: SDRG_14951-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia diclina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia diclina | |||||||||||
| CAZyme ID | SDRG_14951-t26_1-p1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.53:1 | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA15 | 20 | 198 | 5e-53 | 0.9948186528497409 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397269 | LPMO_10 | 3.91e-14 | 20 | 198 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| 395595 | CBM_1 | 1.11e-09 | 301 | 329 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 1.39e-08 | 300 | 333 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 373271 | NSP2_middle | 6.77e-05 | 206 | 266 | 29 | 93 | Middle region of RNA-arterivirus nonstructural protein 2 (nsp2). This domain represents the middle region of nsp2 of the RNA-arteriviruses, such as porcine arterivirus PRRSV and equine arterivirus EAV, C-terminal to the peptidase C33 family catalytic domain. |
| 225689 | DedD | 4.96e-04 | 210 | 296 | 58 | 151 | Cell division protein DedD (periplasmic protein involved in septation) [Cell cycle control, cell division, chromosome partitioning]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.87e-230 | 1 | 338 | 1 | 338 | |
| 1.42e-207 | 1 | 338 | 1 | 371 | |
| 2.94e-134 | 5 | 338 | 3 | 332 | |
| 1.93e-129 | 23 | 336 | 1 | 306 | |
| 7.68e-128 | 1 | 336 | 2 | 342 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.40e-27 | 20 | 201 | 1 | 193 | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State [Thermobia domestica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.83e-08 | 302 | 334 | 22 | 54 | Endo-1,4-beta-xylanase 6 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL6 PE=3 SV=1 |
|
| 5.83e-08 | 302 | 334 | 22 | 54 | Endo-1,4-beta-xylanase 6 OS=Magnaporthe grisea OX=148305 GN=XYL6 PE=1 SV=1 |
|
| 2.98e-07 | 302 | 334 | 32 | 64 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel6A PE=1 SV=1 |
|
| 4.89e-07 | 304 | 334 | 24 | 54 | Probable mannan endo-1,4-beta-mannosidase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manF PE=3 SV=2 |
|
| 5.09e-07 | 292 | 334 | 13 | 55 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
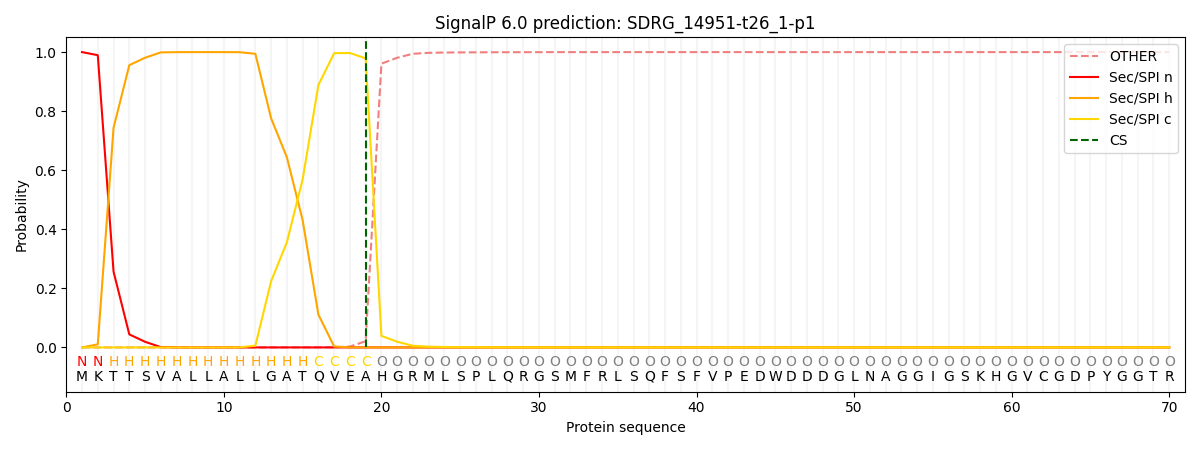
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000297 | 0.999682 | CS pos: 19-20. Pr: 0.9788 |
