You are browsing environment: FUNGIDB
CAZyme Information: SDRG_09930-t26_1-p1
You are here: Home > Sequence: SDRG_09930-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia diclina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia diclina | |||||||||||
| CAZyme ID | SDRG_09930-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 979 | 1706 | 5.6e-274 | 0.9580514208389715 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 979 | 1750 | 3 | 817 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 3.14e-27 | 116 | 210 | 3 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 2 | 1896 | 119 | 2023 | |
| 0.0 | 1 | 1896 | 120 | 2019 | |
| 0.0 | 1 | 1896 | 2152 | 4070 | |
| 3.36e-270 | 25 | 1692 | 69 | 1577 | |
| 5.63e-267 | 25 | 1692 | 92 | 1589 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.96e-260 | 22 | 1875 | 239 | 1917 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 6.60e-251 | 22 | 1883 | 262 | 1904 | Callose synthase 10 OS=Arabidopsis thaliana OX=3702 GN=CALS10 PE=2 SV=5 |
|
| 1.58e-250 | 22 | 1877 | 255 | 1924 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 2.63e-249 | 22 | 1877 | 232 | 1940 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
|
| 1.41e-248 | 3 | 1875 | 228 | 1910 | Putative callose synthase 6 OS=Arabidopsis thaliana OX=3702 GN=CALS6 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
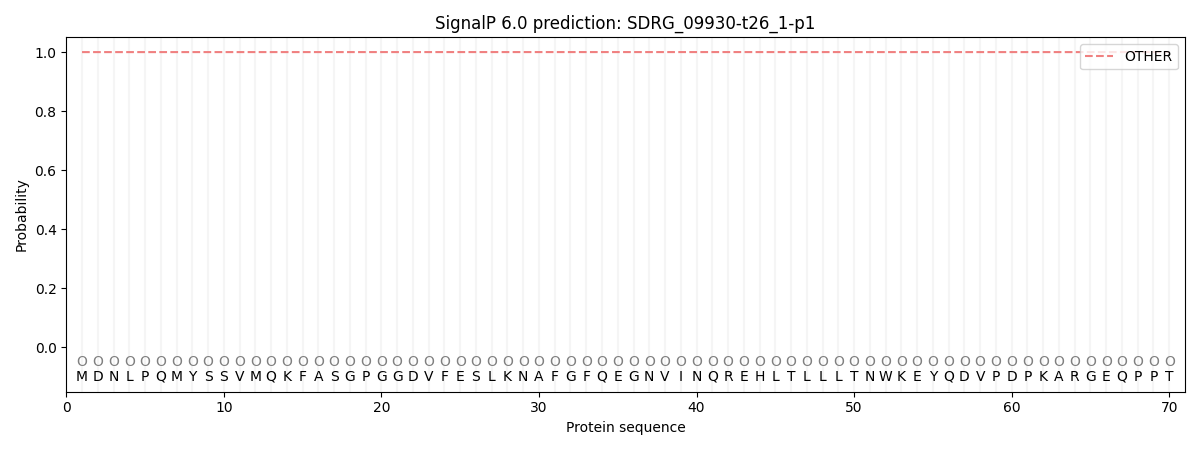
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000000 |
TMHMM Annotations download full data without filtering help
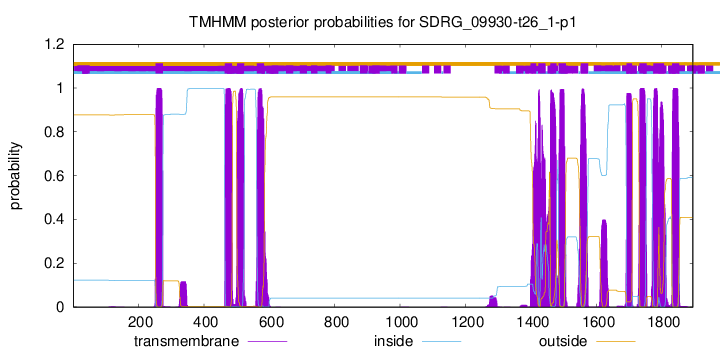
| Start | End |
|---|---|
| 252 | 274 |
| 464 | 486 |
| 501 | 523 |
| 562 | 584 |
| 1399 | 1416 |
| 1423 | 1445 |
| 1460 | 1478 |
| 1485 | 1507 |
| 1553 | 1575 |
| 1693 | 1710 |
| 1732 | 1754 |
| 1775 | 1808 |
| 1832 | 1854 |
