You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS8774-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS8774-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS8774-t41_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 22 | 600 | 7.4e-75 | 0.5097087378640777 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235000 | PRK02106 | 2.50e-64 | 22 | 594 | 2 | 529 | choline dehydrogenase; Validated |
| 225186 | BetA | 3.94e-60 | 19 | 599 | 1 | 536 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 398739 | GMC_oxred_C | 1.59e-25 | 455 | 592 | 4 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 366272 | GMC_oxred_N | 2.22e-25 | 119 | 358 | 19 | 207 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 215420 | PLN02785 | 3.98e-20 | 10 | 588 | 40 | 568 | Protein HOTHEAD |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 5 | 609 | 36 | 651 | |
| 0.0 | 5 | 609 | 36 | 651 | |
| 7.70e-195 | 6 | 589 | 13 | 590 | |
| 4.27e-169 | 10 | 599 | 20 | 667 | |
| 1.71e-168 | 10 | 599 | 20 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.79e-36 | 25 | 604 | 13 | 533 | Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_B Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_C Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_D Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_E Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_F Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_G Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_H Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis] |
|
| 2.79e-36 | 25 | 604 | 13 | 533 | Chain A, Choline oxidase [Arthrobacter globiformis],3LJP_B Chain B, Choline oxidase [Arthrobacter globiformis] |
|
| 6.96e-36 | 25 | 604 | 13 | 533 | Crystal structure of choline oxidase reveals insights into the catalytic mechanism [Arthrobacter globiformis],2JBV_B Crystal structure of choline oxidase reveals insights into the catalytic mechanism [Arthrobacter globiformis],4MJW_A Crystal Structure of Choline Oxidase in Complex with the Reaction Product Glycine Betaine [Arthrobacter globiformis],4MJW_B Crystal Structure of Choline Oxidase in Complex with the Reaction Product Glycine Betaine [Arthrobacter globiformis] |
|
| 3.32e-30 | 26 | 599 | 6 | 530 | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state [Methylovorus sp. MP688],4UDP_B Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state [Methylovorus sp. MP688],4UDQ_A Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state [Methylovorus sp. MP688],4UDQ_B Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state [Methylovorus sp. MP688] |
|
| 3.32e-30 | 26 | 599 | 6 | 530 | Crystal structure of the V465T mutant of 5-(Hydroxymethyl)furfural Oxidase (HMFO) [Methylovorus sp. MP688],6F97_B Crystal structure of the V465T mutant of 5-(Hydroxymethyl)furfural Oxidase (HMFO) [Methylovorus sp. MP688] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.01e-45 | 24 | 596 | 5 | 535 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas savastanoi pv. phaseolicola (strain 1448A / Race 6) OX=264730 GN=betA PE=3 SV=1 |
|
| 2.76e-45 | 24 | 596 | 5 | 535 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=betA PE=3 SV=1 |
|
| 2.50e-44 | 24 | 596 | 5 | 535 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas syringae pv. tomato (strain ATCC BAA-871 / DC3000) OX=223283 GN=betA PE=3 SV=1 |
|
| 1.29e-43 | 17 | 594 | 34 | 571 | Choline dehydrogenase, mitochondrial OS=Mus musculus OX=10090 GN=Chdh PE=1 SV=1 |
|
| 3.41e-43 | 15 | 594 | 35 | 574 | Choline dehydrogenase, mitochondrial OS=Rattus norvegicus OX=10116 GN=Chdh PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
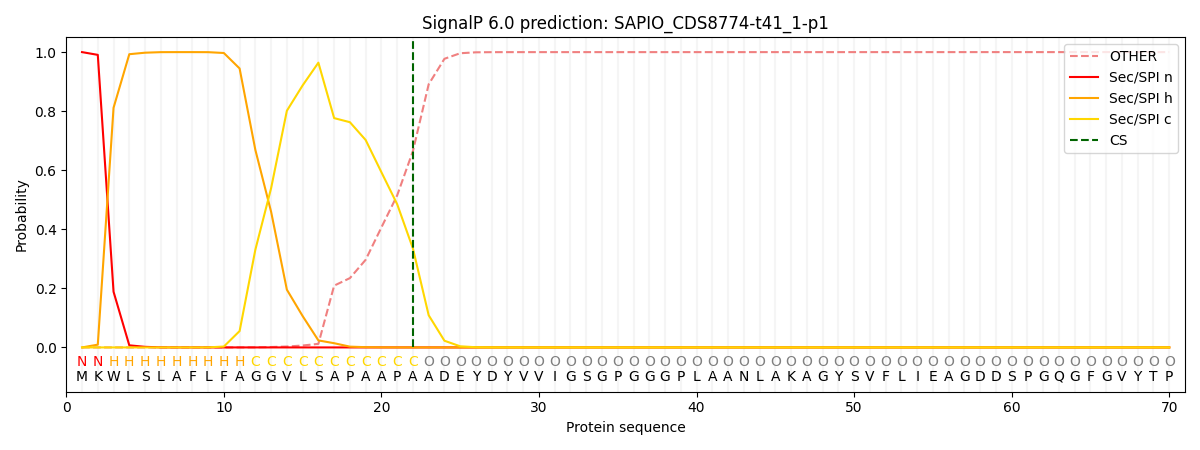
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000358 | 0.999597 | CS pos: 22-23. Pr: 0.3350 |
