You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS3802-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS3802-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS3802-t41_1-p1 | |||||||||||
| CAZy Family | GH114 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 306 | 497 | 2.2e-59 | 0.9948453608247423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398968 | NPP1 | 3.88e-73 | 51 | 255 | 1 | 198 | Necrosis inducing protein (NPP1). This family consists of several NPP1 like necrosis inducing proteins from oomycetes, fungi and bacteria. Infiltration of NPP1 into leaves of Arabidopsis thaliana plants result in transcript accumulation of pathogenesis-related (PR) genes, production of ROS and ethylene, callose apposition, and HR-like cell death. |
| 238871 | XynB_like | 8.16e-49 | 306 | 497 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 404371 | Lipase_GDSL_2 | 2.14e-15 | 311 | 488 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 405210 | HeLo | 7.18e-15 | 1080 | 1206 | 70 | 155 | Prion-inhibition and propagation. This N-terminal region, HeLo, has a prion-inhibitory effect in cis on its own prion-forming domain (PFD) and in trans on HET-s prion propagation. The domain is found exclusively in the fungal kingdom. Its structure, as it occurs in the HET-s/HET-S proteins, consists of two bundles of alpha-helices that pack into a single globular domain. The domain boundary determined from its structure and from protease-resistance experiments overlaps with the C-terminal prion-forming domain of HET-s (PF11558. The HeLo domains of HET-s and HET-S are very similar and their few differences (and not the prion-forming domains) determine the compatibility-phenotype of the fungi in which the proteins are expressed. The mechanism of the HeLo domain-function in heterokaryon-incompatibility is still under investigation, however the HeLo domain is found in similar protein architectures as other cell death and apoptosis-inducing domains. The only other HeLo protein to which a function has been associated is LopB from L. maculans. Although its specific role in L. maculans is unknown, LopB- mutants have impaired ability to form lesions on oilseed rape. The HeLo domain is not related to the HET domain (PF06985) which is another domain involved in heterokaryon incompatibility. |
| 238141 | SGNH_hydrolase | 4.41e-13 | 308 | 495 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.61e-189 | 40 | 836 | 58 | 830 | |
| 1.62e-169 | 10 | 836 | 4 | 827 | |
| 8.51e-161 | 15 | 930 | 8 | 782 | |
| 1.10e-139 | 271 | 1077 | 260 | 1046 | |
| 3.79e-125 | 289 | 1036 | 632 | 1363 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.44e-21 | 55 | 255 | 25 | 210 | Toxin fold for microbial attack and plant defense [Pythium aphanidermatum] |
|
| 2.81e-21 | 55 | 255 | 25 | 210 | NLPPya in complex with glucosamine [Pythium aphanidermatum],5NNW_B NLPPya in complex with glucosamine [Pythium aphanidermatum],5NNW_C NLPPya in complex with glucosamine [Pythium aphanidermatum],5NNW_D NLPPya in complex with glucosamine [Pythium aphanidermatum],5NO9_A NLPPya in complex with mannosamine [Pythium aphanidermatum],5NO9_B NLPPya in complex with mannosamine [Pythium aphanidermatum],5NO9_C NLPPya in complex with mannosamine [Pythium aphanidermatum],5NO9_D NLPPya in complex with mannosamine [Pythium aphanidermatum] |
|
| 8.30e-21 | 55 | 255 | 25 | 210 | Toxin fold as basis for microbial attack and plant defense [Pythium aphanidermatum] |
|
| 8.19e-19 | 55 | 255 | 25 | 210 | Crystal structure of NLPPya P41A, D44N, N48E mutant [Pythium aphanidermatum],6QBD_B Crystal structure of NLPPya P41A, D44N, N48E mutant [Pythium aphanidermatum] |
|
| 4.30e-15 | 41 | 185 | 10 | 149 | Chain A, Necrosis-and ethylene-inducing protein [Moniliophthora perniciosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.78e-21 | 43 | 255 | 33 | 231 | NLP effector protein 10 OS=Phytophthora capsici OX=4784 GN=NLP10 PE=2 SV=1 |
|
| 1.47e-16 | 306 | 498 | 44 | 261 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
|
| 3.48e-16 | 34 | 255 | 26 | 231 | NLP effector protein 1 OS=Phytophthora capsici OX=4784 GN=NLP1 PE=2 SV=1 |
|
| 4.41e-16 | 43 | 257 | 45 | 245 | NLP effector protein 2 OS=Phytophthora capsici OX=4784 GN=NLP2 PE=2 SV=1 |
|
| 4.10e-15 | 43 | 257 | 29 | 235 | NLP effector protein 3 OS=Phytophthora capsici OX=4784 GN=NLP3 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
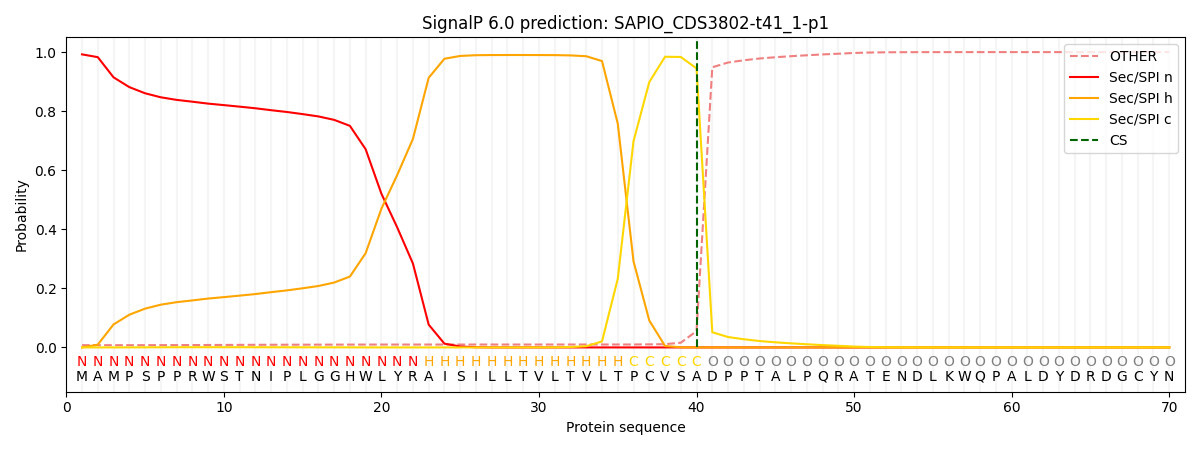
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.008745 | 0.991227 | CS pos: 40-41. Pr: 0.9448 |
