You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS1226-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS1226-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS1226-t41_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 47 | 217 | 6.8e-65 | 0.96045197740113 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 4.48e-90 | 47 | 213 | 2 | 168 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.67e-124 | 1 | 230 | 1 | 222 | |
| 7.02e-115 | 1 | 229 | 1 | 213 | |
| 7.02e-115 | 1 | 229 | 1 | 213 | |
| 7.02e-115 | 1 | 229 | 1 | 213 | |
| 1.56e-111 | 1 | 230 | 1 | 224 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.17e-63 | 40 | 229 | 6 | 189 | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) [Talaromyces funiculosus] |
|
| 1.67e-63 | 47 | 229 | 12 | 186 | 3D Structure of a thermophilic family GH11 xylanase from Thermobifida fusca [Thermobifida fusca] |
|
| 4.69e-63 | 47 | 229 | 55 | 230 | Crystal structure of XlnB2 [Streptomyces lividans],5EJ3_B Crystal structure of XlnB2 [Streptomyces lividans] |
|
| 5.21e-63 | 37 | 229 | 5 | 192 | High resolution structure of GH11 xylanase from Nectria haematococca [Fusarium vanettenii 77-13-4] |
|
| 7.87e-63 | 47 | 229 | 16 | 190 | Chain A, Endo-1,4-beta-xylanase [Actinomycetia bacterium],7EO6_B Chain B, Endo-1,4-beta-xylanase [Actinomycetia bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.29e-73 | 1 | 230 | 1 | 225 | Endo-1,4-beta-xylanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xlnB PE=3 SV=2 |
|
| 2.63e-72 | 5 | 229 | 6 | 224 | Endo-1,4-beta-xylanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xlnA PE=1 SV=1 |
|
| 2.45e-70 | 1 | 230 | 1 | 225 | Probable endo-1,4-beta-xylanase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnB PE=3 SV=1 |
|
| 2.45e-70 | 1 | 230 | 1 | 225 | Endo-1,4-beta-xylanase B OS=Aspergillus niger OX=5061 GN=xlnB PE=1 SV=1 |
|
| 7.69e-70 | 1 | 229 | 1 | 227 | Probable endo-1,4-beta-xylanase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xlnA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
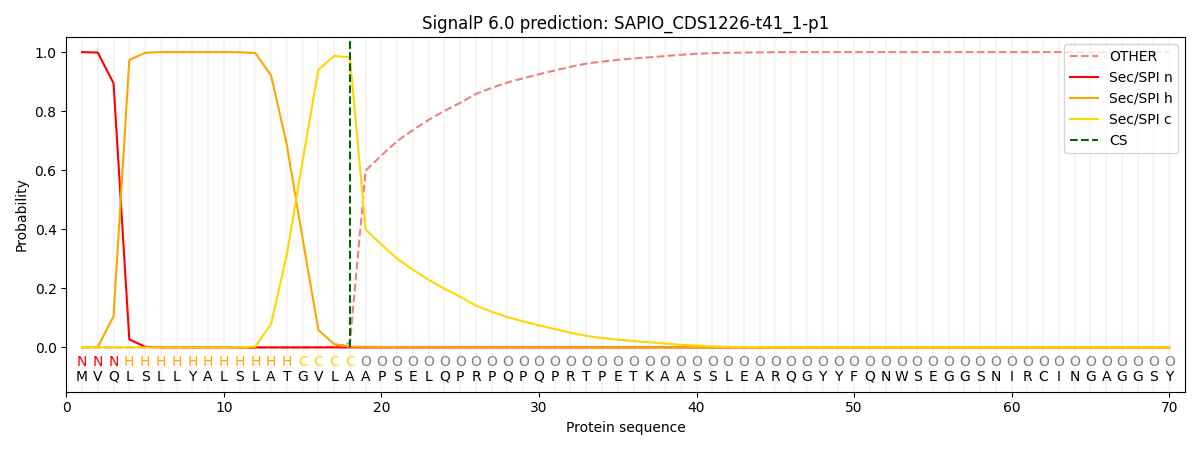
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000269 | 0.999703 | CS pos: 18-19. Pr: 0.9823 |
