You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS1160-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS1160-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
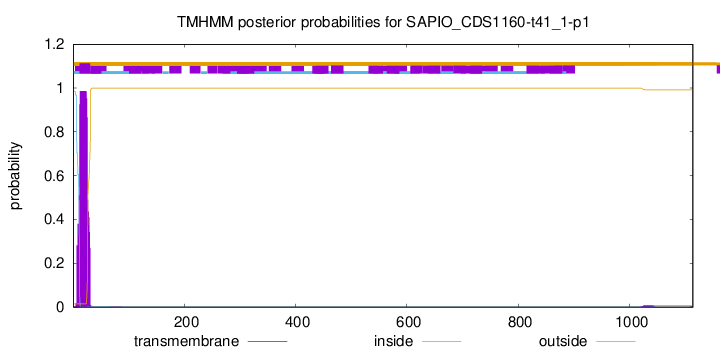
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS1160-t41_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT25 | 52 | 313 | 7.2e-27 | 0.9502762430939227 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173788 | Peptidases_S53 | 4.66e-87 | 622 | 1107 | 1 | 361 | Peptidase domain in the S53 family. Members of the peptidases S53 (sedolisin) family include endopeptidases and exopeptidases sedolisin, kumamolysin, and (PSCP) Pepstatin-insensitive Carboxyl Proteinase. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of Asn in subtilisin. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. Characterized sedolisins include Kumamolisin, an extracellular calcium-dependent thermostable endopeptidase from Bacillus. The enzyme is synthesized with a 188 amino acid N-terminal preprotein region which is cleaved after the extraction into the extracellular space with low pH. One kumamolysin paralog, kumamolisin-As, is believed to be a collagenase. TPP1 is a serine protease that functions as a tripeptidyl exopeptidase as well as an endopeptidase. Less is known about PSCP from Pseudomonas which is thought to be an aspartic proteinase. |
| 401284 | Pro-kuma_activ | 7.80e-37 | 424 | 531 | 37 | 142 | Pro-kumamolisin, activation domain. Members of this family are found in various subtilase propeptides, and adopt a ferredoxin-like fold, with an alpha+beta sandwich. Cleavage of the domain results in activation of the peptide. |
| 206778 | Pro-peptidase_S53 | 3.14e-30 | 424 | 527 | 36 | 138 | Activation domain of S53 peptidases. Members of this family are found in various subtilase propeptides, such as pro-kumamolysin and tripeptidyl peptidase I, and adopt a ferredoxin-like fold, with an alpha+beta sandwich. Cleavage of the domain results in activation of the peptidase. |
| 133474 | Glyco_transf_25 | 1.57e-26 | 52 | 141 | 1 | 96 | Glycosyltransferase family 25 [lipooligosaccharide (LOS) biosynthesis protein] is a family of glycosyltransferases involved in LOS biosynthesis. The members include the beta(1,4) galactosyltransferases: Lgt2 of Moraxella catarrhalis, LgtB and LgtE of Neisseria gonorrhoeae and Lic2A of Haemophilus influenzae. M. catarrhalis Lgt2 catalyzes the addition of galactose (Gal) to the growing chain of LOS on the cell surface. N. gonorrhoeae LgtB and LgtE link Gal-beta(1,4) to GlcNAc (N-acetylglucosamine) and Glc (glucose), respectively. The genes encoding LgtB and LgtE are two genes of a five gene locus involved in the synthesis of gonococcal LOS. LgtE is believed to perform the first step in LOS biosynthesis. |
| 214928 | Pro-kuma_activ | 4.09e-25 | 424 | 527 | 34 | 134 | Pro-kumamolisin, activation domain. This domain is found at the N-terminus of peptidases belonging to MEROPS peptidase family S53 (sedolisin, clan SB). The domain adopts a ferredoxin-like fold, with an alpha+beta sandwich. Cleavage of the domain results in activation of the peptidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.16e-103 | 28 | 403 | 32 | 397 | |
| 2.00e-99 | 10 | 400 | 5 | 391 | |
| 4.63e-95 | 42 | 403 | 36 | 393 | |
| 2.50e-94 | 28 | 401 | 32 | 387 | |
| 2.50e-94 | 28 | 401 | 32 | 387 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.43e-35 | 424 | 1112 | 50 | 543 | Crystal Structure of the Precursor Form of Human Tripeptidyl-Peptidase 1 [Homo sapiens] |
|
| 1.02e-34 | 424 | 1112 | 69 | 562 | Crystal Structure Analysis of Tripeptidyl peptidase -I [Homo sapiens],3EE6_B Crystal Structure Analysis of Tripeptidyl peptidase -I [Homo sapiens] |
|
| 2.93e-16 | 770 | 1111 | 101 | 356 | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) [Bacillus subtilis],1GT9_2 High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) [Bacillus subtilis],1GTG_1 Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) [Bacillus sp. MN-32],1GTJ_1 Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho [Bacillus sp. MN-32],1GTJ_2 Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho [Bacillus sp. MN-32],1GTL_1 The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho [Bacillus sp. MN-32],1GTL_2 The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho [Bacillus sp. MN-32] |
|
| 3.16e-16 | 770 | 1111 | 101 | 356 | Chain A, kumamolisin [Bacillus sp. MN-32] |
|
| 1.01e-15 | 770 | 1111 | 101 | 356 | Chain A, kumamolisin [Bacillus sp. MN-32] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.59e-148 | 418 | 1114 | 65 | 644 | Tripeptidyl-peptidase sed1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=sed1 PE=1 SV=1 |
|
| 3.60e-121 | 424 | 1114 | 72 | 652 | Aorsin OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=aorO PE=1 SV=2 |
|
| 6.34e-74 | 412 | 1113 | 63 | 669 | Tripeptidyl-peptidase SED1 OS=Arthroderma otae (strain ATCC MYA-4605 / CBS 113480) OX=554155 GN=SED1 PE=3 SV=1 |
|
| 4.29e-41 | 420 | 1112 | 61 | 557 | Tripeptidyl-peptidase 1 OS=Danio rerio OX=7955 GN=tpp1 PE=1 SV=2 |
|
| 1.04e-39 | 424 | 1109 | 71 | 594 | Tripeptidyl-peptidase SED2 OS=Arthroderma otae (strain ATCC MYA-4605 / CBS 113480) OX=554155 GN=SED2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999082 | 0.000933 |