You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS10548-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS10548-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS10548-t41_1-p1 | |||||||||||
| CAZy Family | AA6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 28 | 468 | 1.2e-194 | 0.9932279909706546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227807 | XynC | 1.67e-26 | 27 | 468 | 33 | 427 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| 405300 | Glyco_hydr_30_2 | 1.14e-17 | 29 | 339 | 3 | 354 | O-Glycosyl hydrolase family 30. |
| 407314 | Glyco_hydro_30C | 6.00e-09 | 379 | 436 | 1 | 62 | Glycosyl hydrolase family 30 beta sandwich domain. |
| 307945 | Glyco_hydro_30 | 1.91e-08 | 43 | 241 | 3 | 197 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| 396577 | Glyco_hydro_59 | 5.31e-07 | 60 | 341 | 20 | 256 | Glycosyl hydrolase family 59. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.59e-238 | 17 | 472 | 16 | 480 | |
| 2.53e-221 | 17 | 472 | 16 | 462 | |
| 2.53e-221 | 17 | 472 | 16 | 462 | |
| 2.07e-186 | 19 | 472 | 17 | 474 | |
| 2.07e-186 | 19 | 472 | 17 | 474 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.27e-185 | 28 | 472 | 3 | 450 | Chain A, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 3.23e-185 | 20 | 472 | 1 | 457 | Chain AAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 5.48e-133 | 30 | 468 | 24 | 470 | Chain A, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
|
| 2.58e-132 | 21 | 468 | 81 | 541 | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
|
| 9.03e-110 | 30 | 469 | 25 | 453 | Chain A, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.68e-187 | 19 | 472 | 17 | 474 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
|
| 2.60e-11 | 28 | 420 | 86 | 495 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
|
| 2.34e-07 | 140 | 469 | 112 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
|
| 4.85e-07 | 22 | 240 | 83 | 298 | Putative glucosylceramidase 3 OS=Caenorhabditis elegans OX=6239 GN=gba-3 PE=3 SV=1 |
|
| 9.65e-07 | 32 | 418 | 31 | 378 | Galactocerebrosidase OS=Salmo salar OX=8030 GN=galc PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
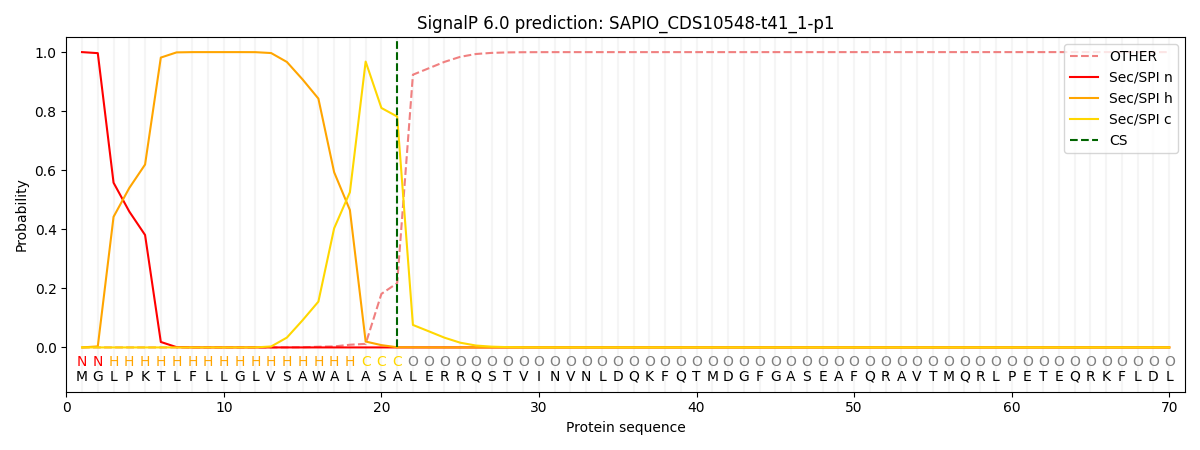
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000253 | 0.999706 | CS pos: 21-22. Pr: 0.7818 |
