You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS0862-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS0862-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
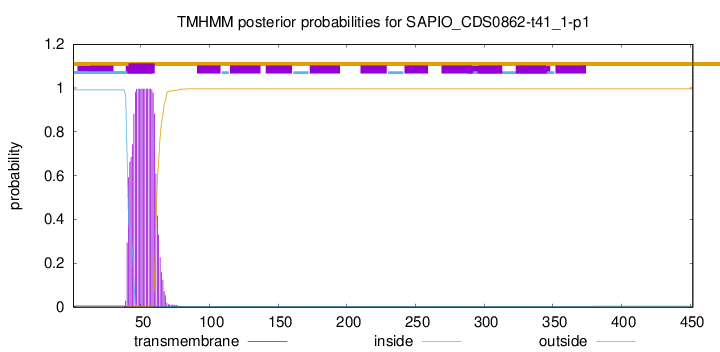
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS0862-t41_1-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 83 | 346 | 1.1e-122 | 0.9888059701492538 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 1.07e-156 | 84 | 346 | 1 | 265 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 404741 | DUF4207 | 0.001 | 387 | 449 | 97 | 152 | Domain of unknown function (DUF4207). This family is found in eukaryotes; it has several conserved tryptophan residues. The function is not known. |
| 406200 | DUF4670 | 0.003 | 389 | 447 | 345 | 404 | Domain of unknown function (DUF4670). This family of proteins is found in eukaryotes. Proteins in this family are typically between 373 and 763 amino acids in length. |
| 406371 | Casc1_N | 0.008 | 390 | 437 | 4 | 44 | Cancer susceptibility candidate 1 N-terminus. This presumed domain is functionally uncharacterized. This domain family is found in eukaryotes, and is approximately 200 amino acids in length. The family is found in association with pfam12366. There are two completely conserved residues (N and W) that may be functionally important. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.15e-265 | 1 | 439 | 2 | 440 | |
| 1.79e-265 | 1 | 439 | 14 | 452 | |
| 8.50e-265 | 1 | 439 | 14 | 452 | |
| 2.34e-264 | 1 | 439 | 2 | 440 | |
| 2.97e-264 | 1 | 435 | 13 | 447 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.50e-62 | 85 | 382 | 6 | 304 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-163 | 16 | 399 | 11 | 393 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 3.16e-162 | 84 | 431 | 61 | 408 | Mannan polymerase II complex ANP1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ANP1 PE=1 SV=3 |
|
| 1.04e-126 | 84 | 396 | 171 | 530 | Mannan polymerase I complex VAN1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=VAN1 PE=1 SV=3 |
|
| 4.21e-122 | 79 | 396 | 70 | 437 | Vanadate resistance protein OS=Candida albicans OX=5476 GN=VAN1 PE=3 SV=1 |
|
| 2.70e-61 | 85 | 380 | 73 | 368 | Mannan polymerase complex subunit MNN9 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN9 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999971 | 0.000049 |