You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS0439-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS0439-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS0439-t41_1-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Xyloglucan-specific exo-beta-1,4-glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:31 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 26 | 332 | 1.4e-96 | 0.966996699669967 |
| CBM1 | 376 | 404 | 1.7e-16 | 0.9655172413793104 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 2.65e-138 | 21 | 332 | 1 | 308 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 5.61e-113 | 67 | 332 | 2 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 2.91e-78 | 7 | 340 | 5 | 345 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| 395595 | CBM_1 | 4.37e-16 | 376 | 404 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 5.41e-16 | 376 | 408 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.79e-202 | 19 | 408 | 18 | 408 | |
| 5.82e-190 | 1 | 408 | 1 | 414 | |
| 1.56e-188 | 1 | 408 | 1 | 418 | |
| 1.56e-188 | 1 | 408 | 1 | 418 | |
| 7.47e-188 | 1 | 408 | 1 | 413 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.13e-132 | 18 | 408 | 22 | 406 | GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8M_B GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8N_A GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872],6Q8N_B GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872] |
|
| 2.84e-118 | 19 | 338 | 1 | 319 | Crystal structure of GH10 family xylanase XynAF1 from Aspergillus fumigatus Z5 [Aspergillus fumigatus Z5],6JDT_B Crystal structure of GH10 family xylanase XynAF1 from Aspergillus fumigatus Z5 [Aspergillus fumigatus Z5],6JDY_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 120 minutes [Aspergillus fumigatus Z5],6JDY_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 120 minutes [Aspergillus fumigatus Z5],6JDZ_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 20 minutes [Aspergillus fumigatus Z5],6JDZ_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 20 minutes [Aspergillus fumigatus Z5],6JE0_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 30 minutes [Aspergillus fumigatus Z5],6JE0_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 30 minutes [Aspergillus fumigatus Z5],6JE1_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 40 minutes [Aspergillus fumigatus Z5],6JE1_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 40 minutes [Aspergillus fumigatus Z5],6JE2_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 80 minutes [Aspergillus fumigatus Z5],6JE2_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 80 minutes [Aspergillus fumigatus Z5] |
|
| 2.22e-111 | 17 | 336 | 1 | 324 | A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft [Fusarium oxysporum],3U7B_B A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft [Fusarium oxysporum],3U7B_C A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft [Fusarium oxysporum],3U7B_D A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft [Fusarium oxysporum],3U7B_E A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft [Fusarium oxysporum] |
|
| 6.33e-92 | 19 | 336 | 3 | 312 | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 1.56e-89 | 19 | 336 | 3 | 312 | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.12e-186 | 3 | 408 | 4 | 429 | Endo-1,4-beta-xylanase 1 OS=Humicola grisea var. thermoidea OX=5528 GN=xyn1 PE=2 SV=1 |
|
| 6.98e-133 | 21 | 344 | 38 | 361 | Endo-1,4-beta-xylanase 5 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL5 PE=3 SV=1 |
|
| 6.98e-133 | 21 | 344 | 38 | 361 | Endo-1,4-beta-xylanase 5 OS=Magnaporthe grisea OX=148305 GN=XYL5 PE=3 SV=1 |
|
| 4.53e-126 | 1 | 408 | 3 | 407 | Endo-1,4-beta-xylanase D OS=Talaromyces funiculosus OX=28572 GN=xynD PE=1 SV=1 |
|
| 2.70e-116 | 27 | 338 | 27 | 342 | Endo-1,4-beta-xylanase 1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYL1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
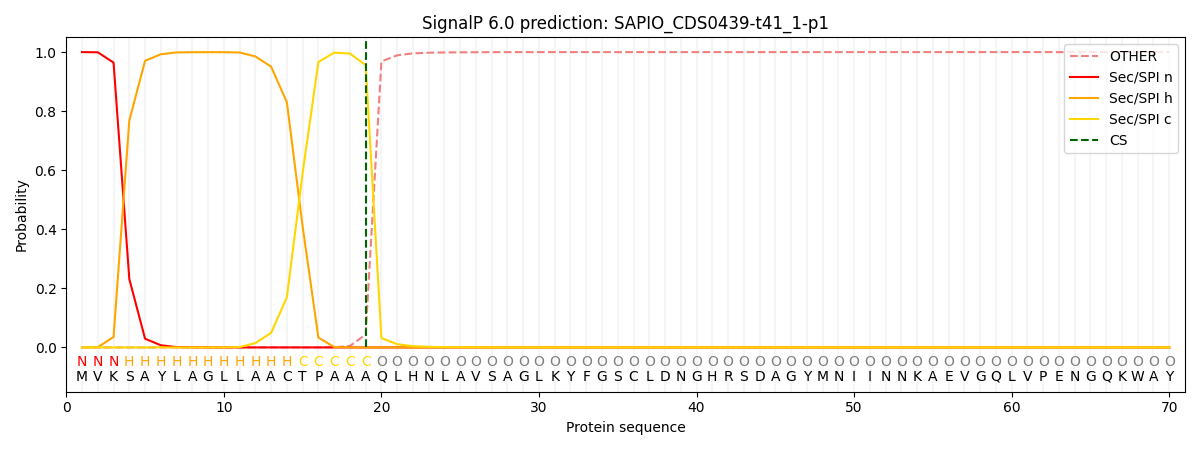
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000360 | 0.999614 | CS pos: 19-20. Pr: 0.9563 |
