You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS0410-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS0410-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS0410-t41_1-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | Cellulase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.91:55 | 3.2.1.4:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 139 | 422 | 1.2e-90 | 0.935374149659864 |
| CBM1 | 32 | 59 | 9.9e-16 | 0.9310344827586207 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396075 | Glyco_hydro_6 | 8.33e-159 | 140 | 426 | 1 | 293 | Glycosyl hydrolases family 6. |
| 227616 | CelA1 | 8.38e-44 | 96 | 461 | 8 | 442 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| 395595 | CBM_1 | 7.40e-16 | 32 | 59 | 2 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 8.15e-16 | 30 | 63 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.73e-220 | 11 | 462 | 9 | 482 | |
| 6.14e-219 | 3 | 462 | 2 | 482 | |
| 1.02e-215 | 3 | 462 | 2 | 484 | |
| 4.15e-215 | 3 | 462 | 2 | 484 | |
| 4.15e-215 | 3 | 462 | 2 | 484 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.12e-193 | 108 | 462 | 1 | 364 | Chain A, Cellobiohydrolase Family 6 [Thermochaetoides thermophila] |
|
| 8.10e-179 | 111 | 462 | 1 | 362 | Structure of the wild-type cellobiohydrolase Cel6A from Humicolas insolens in complex with a fluorescent substrate [Humicola insolens],1OCB_B Structure of the wild-type cellobiohydrolase Cel6A from Humicolas insolens in complex with a fluorescent substrate [Humicola insolens],2BVW_A CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE [Humicola insolens],2BVW_B CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE [Humicola insolens] |
|
| 1.52e-178 | 113 | 462 | 1 | 360 | Cellobiohydrolase Ii (Cel6a) From Humicola Insolens [Humicola insolens] |
|
| 1.75e-178 | 109 | 462 | 1 | 364 | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside [Humicola insolens],1OC6_A structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution [Humicola insolens],1OC7_A D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-tetrathio-alpha-d-cellopentoside at 1.1 angstrom resolution [Humicola insolens] |
|
| 1.33e-177 | 111 | 462 | 1 | 362 | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution [Humicola insolens],1OCN_A Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.37e-216 | 3 | 462 | 2 | 484 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=CEL6A PE=1 SV=1 |
|
| 3.11e-208 | 7 | 462 | 4 | 476 | Exoglucanase-6A OS=Humicola insolens OX=34413 GN=cel6A PE=1 SV=1 |
|
| 2.06e-191 | 15 | 462 | 7 | 454 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=cbhC PE=3 SV=1 |
|
| 2.06e-191 | 15 | 462 | 7 | 454 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=cbhC PE=3 SV=1 |
|
| 5.90e-190 | 15 | 462 | 7 | 450 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
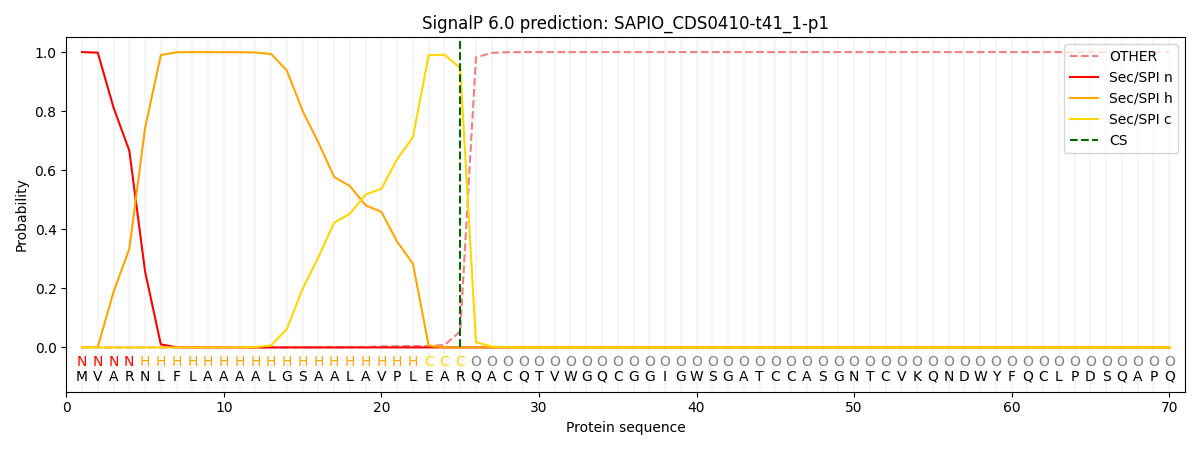
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000327 | 0.999649 | CS pos: 25-26. Pr: 0.9461 |
