You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS0363-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS0363-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS0363-t41_1-p1 | |||||||||||
| CAZy Family | AA16 | |||||||||||
| CAZyme Description | Xyloglucan-specific exo-beta-1,4-glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 182430; End:185328 Strand: + | |||||||||||
Full Sequence Download help
| MKSILLASLA AWGAYAQSGA WGQCGGINWS GPTSCISGYT CVYQNDWYSQ CLPASQVPTT | 60 |
| TTKAPSTTLT TSTRPPTSTS NPGNGSKKMK WVGVNLSVAE FGQGTYPGTW GKEFYFPDNN | 120 |
| AVSTLISQGY NTFRVAFAME RMAVGSLTSN FDQGYLTNLT ATVNHITQNG AWAVLDPHNF | 180 |
| GRYNGNVITD TNAFKAFWVK LATHFKNNDK VIFDTNNEYN TMESSLVFDL NQAAINGIRE | 240 |
| TGATQYIFVE GNSWSGAWTW TQINTNLVSL QDPQNKIVYE MHQYLDSDGS GTSTACVSNT | 300 |
| IGVERVTAAT KWLKDNGKVG FLGEFAGGAN ANCKEAVTGM LNHLQDNSDV WLGALWWAAG | 360 |
| PWWGNYIYSF EPPSGTGYTY YNSLLKQAYP WTSKPLIVLA PMRPVSGPHL AVAVSRAGGL | 420 |
| GFINPNGTHQ GIIDDFEDVK KLLEAQDTSS DALSKFASGG LLPVGIGFIV WRDDKQVALS | 480 |
| TVEKYRPVAV WLFAPRTGQG ELDEWISELR NVSPQTQIWI QVGTLGEATA AANSSSPPDV | 540 |
| LVIQGTEAGG HGRTSDGLSI VTLFPEVANA IRGSGIPLIA TGGIADGRGV AAALSLGAAG | 600 |
| VALGTRFIAA TEARVKKGYQ RAVLDAKDGA KSTVRTTIYN QLRGTNWPEQ YSPRGVTNRT | 660 |
| WDDFQAKVPY EELKKAFDES TKAGDEGWGI EGRQPTYAGA GVGLVDTVAD AETIVNNLRK | 720 |
| DAISIAKAVA ESFE | 734 |
Enzyme Prediction help
| EC | 3.2.1.4:38 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 91 | 367 | 5.3e-109 | 0.9964539007092199 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 9.66e-67 | 105 | 360 | 14 | 272 | Cellulase (glycosyl hydrolase family 5). |
| 240081 | NPD_like | 7.20e-48 | 394 | 644 | 2 | 225 | 2-Nitropropane dioxygenase (NPD), one of the nitroalkane oxidizing enzyme families, catalyzes oxidative denitrification of nitroalkanes to their corresponding carbonyl compounds and nitrites. NDP is a member of the NAD(P)H-dependent flavin oxidoreductase family that reduce a range of alternative electron acceptors. Most use FAD/FMN as a cofactor and NAD(P)H as electron donor. Some contain 4Fe-4S cluster to transfer electron from FAD to FMN. |
| 224981 | YrpB | 3.75e-33 | 397 | 728 | 17 | 331 | NAD(P)H-dependent flavin oxidoreductase YrpB, nitropropane dioxygenase family [General function prediction only]. |
| 367316 | NMO | 1.08e-20 | 392 | 718 | 9 | 328 | Nitronate monooxygenase. Nitronate monooxygenase (NMO), formerly referred to as 2-nitropropane dioxygenase (NPD) (EC:1.13.11.32), is an FMN-dependent enzyme that uses molecular oxygen to oxidize (anionic) alkyl nitronates and, in the case of the enzyme from Neurospora crassa, (neutral) nitroalkanes to the corresponding carbonyl compounds and nitrite. Previously classified as 2-nitropropane dioxygenase, but it is now recognized that this was the result of the slow ionization of nitroalkanes to their nitronate (anionic) forms. The enzymes from the fungus Neurospora crassa and the yeast Williopsis saturnus var. mrakii (formerly classified as Hansenula mrakii) contain non-covalently bound FMN as the cofactor. Active towards linear alkyl nitronates of lengths between 2 and 6 carbon atoms and, with lower activity, towards propyl-2-nitronate. The enzyme from N. crassa can also utilize neutral nitroalkanes, but with lower activity. One atom of oxygen is incorporated into the carbonyl group of the aldehyde product. The reaction appears to involve the formation of an enzyme-bound nitronate radical and an a-peroxynitroethane species, which then decomposes, either in the active site of the enzyme or after release, to acetaldehyde and nitrite. |
| 132195 | enACPred_II | 1.06e-18 | 384 | 628 | 7 | 214 | putative enoyl-[acyl-carrier-protein] reductase II. This oxidoreductase of the 2-nitropropane dioxygenase family (pfam03060) is commonly found in apparent operons with genes involved in fatty acid biosynthesis. Furthermore, this genomic context generally includes the fabG 3-oxoacyl-[ACP] reductase and lacks the fabI enoyl-[ACP] reductase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEO53769.1|CBM1|GH5_5|3.2.1.4 | 1.77e-181 | 1 | 390 | 1 | 389 |
| AEM23898.1|CBM1|GH5_5|3.2.1.4 | 1.77e-181 | 1 | 390 | 1 | 389 |
| CAE81955.1|CBM1|GH5_5|3.2.1.4 | 7.75e-178 | 1 | 390 | 1 | 390 |
| BAA12676.1|CBM1|GH5_5|3.2.1.4 | 9.37e-176 | 1 | 390 | 1 | 388 |
| CAA53631.1|CBM1|GH5_5|3.2.1.4 | 3.03e-174 | 1 | 390 | 1 | 388 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5I77_A | 7.89e-131 | 90 | 379 | 3 | 292 | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger] |
| 5I79_A | 6.72e-130 | 90 | 379 | 5 | 294 | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger],5I79_B Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger] |
| 5I78_A | 6.95e-130 | 90 | 379 | 6 | 295 | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger],5I78_B Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger] |
| 1GZJ_A | 1.87e-126 | 87 | 378 | 1 | 293 | Structure of Thermoascus aurantiacus family 5 endoglucanase [Thermoascus aurantiacus],1GZJ_B Structure of Thermoascus aurantiacus family 5 endoglucanase [Thermoascus aurantiacus] |
| 1H1N_A | 4.31e-125 | 86 | 378 | 1 | 294 | Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus [Thermoascus aurantiacus],1H1N_B Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q7SDR1|GUN3_NEUCR | 1.38e-178 | 1 | 390 | 1 | 390 | Endoglucanase gh5-1 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=gh5-1 PE=1 SV=1 |
| sp|Q12624|GUN3_HUMIN | 5.38e-175 | 1 | 390 | 1 | 388 | Endoglucanase 3 OS=Humicola insolens OX=34413 GN=CMC3 PE=1 SV=1 |
| sp|A1DME8|EGLB_NEOFI | 5.21e-139 | 82 | 378 | 20 | 317 | Probable endo-beta-1,4-glucanase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglB PE=3 SV=1 |
| sp|O74706|EGLB_ASPNG | 9.96e-131 | 83 | 379 | 22 | 321 | Endo-beta-1,4-glucanase B OS=Aspergillus niger OX=5061 GN=eglB PE=1 SV=1 |
| sp|A2QPC3|EGLB_ASPNC | 9.96e-131 | 83 | 379 | 22 | 321 | Probable endo-beta-1,4-glucanase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
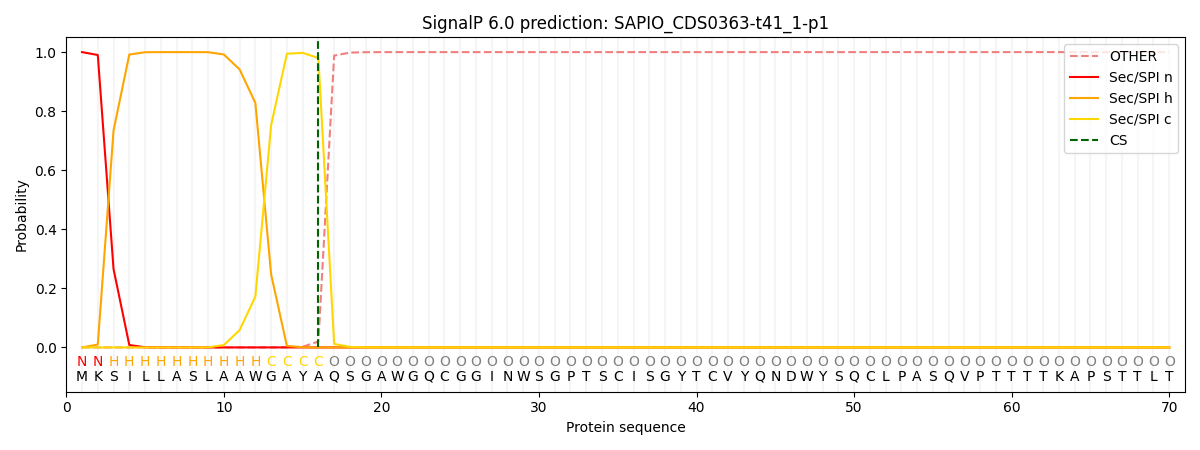
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000276 | 0.999698 | CS pos: 16-17. Pr: 0.9800 |
