You are browsing environment: FUNGIDB
CAZyme Information: SAPIO_CDS0306-t41_1-p1
You are here: Home > Sequence: SAPIO_CDS0306-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scedosporium apiospermum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Scedosporium; Scedosporium apiospermum | |||||||||||
| CAZyme ID | SAPIO_CDS0306-t41_1-p1 | |||||||||||
| CAZy Family | AA12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.99.18:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA8 | 26 | 210 | 2.2e-64 | 0.9945054945054945 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 406418 | CDH-cyt | 1.21e-68 | 26 | 206 | 1 | 177 | Cytochrome domain of cellobiose dehydrogenase. CDH-cyt is the cytochrome domain, at the N-terminus, of cellobiose dehydrogenase. CDH-cyt folds as a beta sandwich with the topology of the antibody Fab V(H) domain and binds iron. The haem iron is ligated by Met83 and His181 in UniProtKB:Q01738. |
| 187688 | CDH_like_cytochrome | 4.31e-48 | 25 | 198 | 1 | 168 | Heme-binding cytochrome domain of fungal cellobiose dehydrogenases. Cellobiose dehydrogenase (CellobioseDH or CDH) is an extracellular fungal oxidoreductase that degrades both lignin and cellulose. Specifically, CDHs oxidize cellobiose, cellodextrins, and lactose to corresponding lactones, utilizing a variety of electron acceptors. Class-II CDHs are monomeric hemoflavoenzymes that are comprised of a b-type cytochrome domain linked to a large flavodehydrogenase domain. The cytochrome domain of CDH and related enzymes, which this model describes, folds as a beta sandwich and complexes a heme molecule. It is found at the N-terminus of this family of enzymes, and belongs to the DOMON domain superfamily, a ligand-interacting motif found in all three kingdoms of life. |
| 197609 | LysM | 1.07e-08 | 248 | 291 | 1 | 44 | Lysin motif. |
| 396179 | LysM | 1.03e-07 | 249 | 292 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| 212030 | LysM | 2.95e-07 | 248 | 291 | 2 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.52e-72 | 8 | 222 | 11 | 237 | |
| 1.69e-66 | 13 | 224 | 173 | 392 | |
| 1.89e-66 | 8 | 249 | 9 | 257 | |
| 2.00e-66 | 20 | 224 | 23 | 235 | |
| 3.90e-66 | 20 | 224 | 77 | 289 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.87e-58 | 26 | 219 | 8 | 208 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI3_B Chain B, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 3.34e-55 | 26 | 224 | 8 | 213 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 1.28e-50 | 26 | 224 | 7 | 213 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
|
| 5.20e-26 | 26 | 177 | 6 | 153 | Chain A, CELLOBIOSE DEHYDROGENASE [Phanerodontia chrysosporium],1D7B_B Chain B, CELLOBIOSE DEHYDROGENASE [Phanerodontia chrysosporium],1D7D_A Chain A, CELLOBIOSE DEHYDROGENASE [Phanerodontia chrysosporium],1D7D_B Chain B, CELLOBIOSE DEHYDROGENASE [Phanerodontia chrysosporium] |
|
| 5.76e-26 | 26 | 177 | 6 | 153 | Chain A, CELLOBIOSE DEHYDROGENASE [Phanerodontia chrysosporium],1D7C_B Chain B, CELLOBIOSE DEHYDROGENASE [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.28e-25 | 8 | 177 | 6 | 171 | Cellobiose dehydrogenase OS=Phanerodontia chrysosporium OX=2822231 GN=CDH-1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
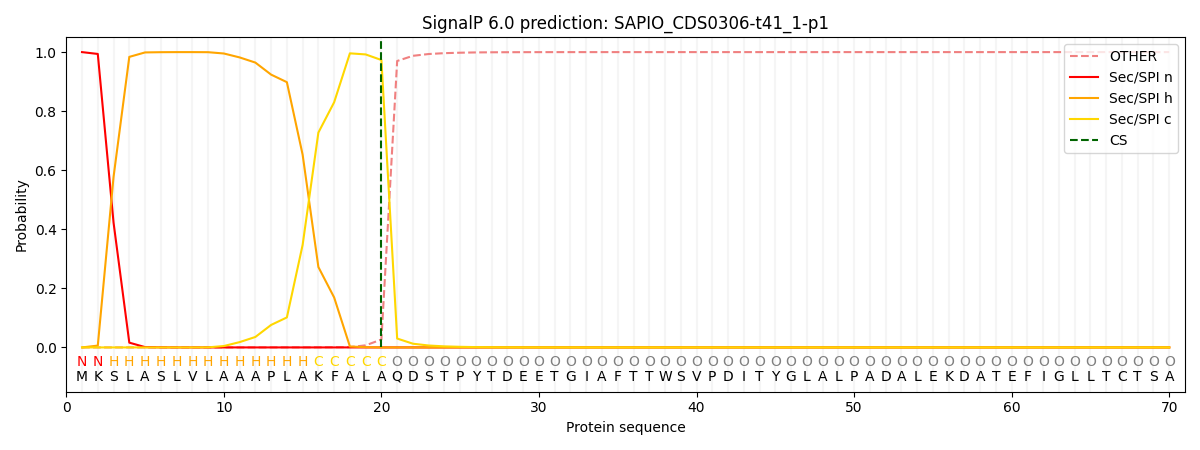
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000238 | 0.999744 | CS pos: 20-21. Pr: 0.9732 |
