You are browsing environment: FUNGIDB
CAZyme Information: RhiirA1_535620-t46_1-p1
You are here: Home > Sequence: RhiirA1_535620-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | RhiirA1_535620-t46_1-p1 | |||||||||||
| CAZy Family | GT57 | |||||||||||
| CAZyme Description | UDP-Glycosyltransferase/glycogen phosphorylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 245 | 475 | 9.1e-33 | 0.5130890052356021 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 7.86e-53 | 40 | 476 | 2 | 403 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 278624 | UDPGT | 1.98e-34 | 51 | 529 | 11 | 483 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 224732 | YjiC | 1.10e-23 | 41 | 475 | 4 | 396 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 223071 | egt | 2.98e-22 | 244 | 487 | 255 | 474 | ecdysteroid UDP-glucosyltransferase; Provisional |
| 178032 | PLN02410 | 2.35e-10 | 49 | 455 | 17 | 419 | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.41e-59 | 39 | 512 | 10 | 502 | |
| 1.00e-58 | 39 | 513 | 9 | 502 | |
| 1.00e-58 | 39 | 513 | 9 | 502 | |
| 1.00e-58 | 39 | 513 | 9 | 502 | |
| 2.64e-58 | 39 | 513 | 9 | 502 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.94e-12 | 266 | 452 | 19 | 182 | Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_B Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_C Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_D Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens] |
|
| 7.78e-12 | 366 | 452 | 75 | 161 | Chain A, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_B Chain B, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_C Chain C, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_D Chain D, UDP-glucuronosyltransferase 2B15 [Homo sapiens] |
|
| 6.17e-10 | 270 | 452 | 1 | 160 | Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens],2O6L_B Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens] |
|
| 1.51e-09 | 49 | 475 | 22 | 429 | Structural Characterization of UDP-glycosyltransferase from Tetranychus Urticae [Tetranychus urticae] |
|
| 1.79e-08 | 361 | 475 | 284 | 395 | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A [Streptomyces antibioticus],4M83_B Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A [Streptomyces antibioticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.02e-24 | 51 | 529 | 37 | 507 | UDP-glucuronosyltransferase 1-6 OS=Homo sapiens OX=9606 GN=UGT1A6 PE=1 SV=2 |
|
| 1.09e-23 | 193 | 529 | 203 | 509 | UDP-glucuronosyltransferase 1A3 OS=Homo sapiens OX=9606 GN=UGT1A3 PE=1 SV=1 |
|
| 1.44e-23 | 245 | 529 | 253 | 506 | UDP-glucuronosyltransferase 1A7 OS=Mus musculus OX=10090 GN=Ugt1a7 PE=1 SV=1 |
|
| 1.96e-23 | 250 | 529 | 261 | 509 | UDP-glucuronosyltransferase 1A4 OS=Homo sapiens OX=9606 GN=UGT1A4 PE=1 SV=1 |
|
| 1.96e-23 | 250 | 529 | 261 | 509 | UDP-glucuronosyltransferase 1A5 OS=Homo sapiens OX=9606 GN=UGT1A5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
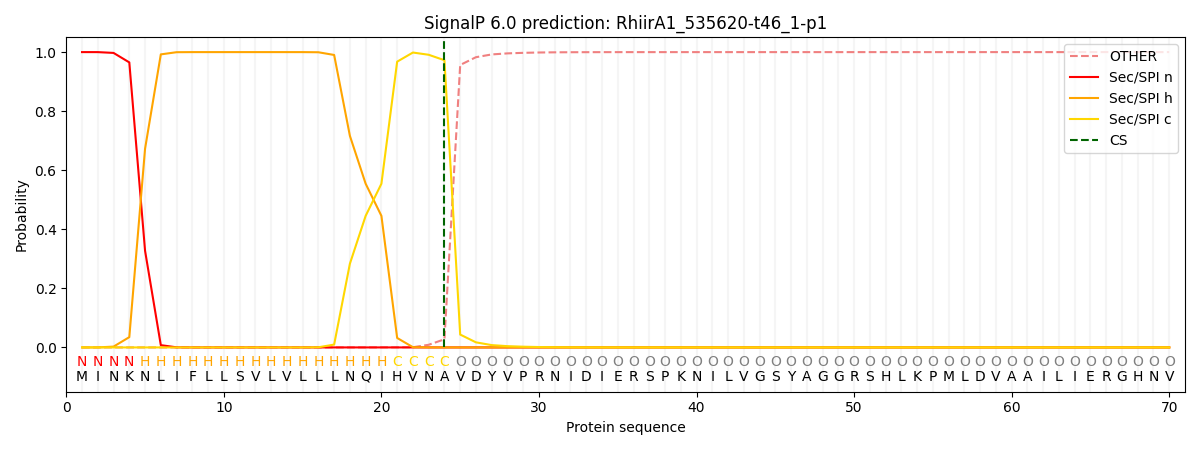
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000205 | 0.999762 | CS pos: 24-25. Pr: 0.9725 |

