You are browsing environment: FUNGIDB
CAZyme Information: RhiirA1_491244-t46_1-p1
You are here: Home > Sequence: RhiirA1_491244-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
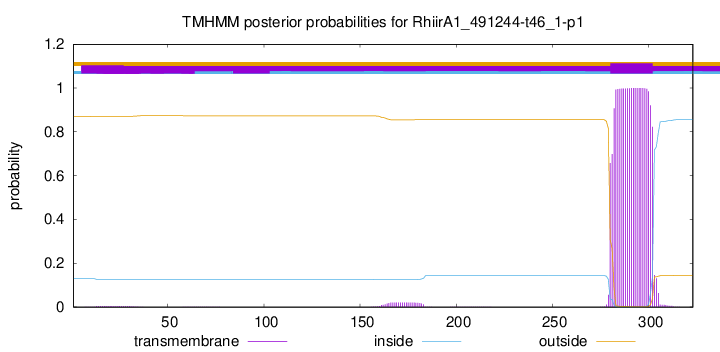
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | RhiirA1_491244-t46_1-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | UDP-Glycosyltransferase/glycogen phosphorylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 46 | 233 | 1.5e-25 | 0.39267015706806285 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 7.11e-32 | 2 | 252 | 177 | 404 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 223071 | egt | 5.04e-21 | 34 | 232 | 264 | 443 | ecdysteroid UDP-glucosyltransferase; Provisional |
| 278624 | UDPGT | 5.42e-21 | 21 | 303 | 231 | 483 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 224732 | YjiC | 5.50e-19 | 33 | 250 | 211 | 396 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 340818 | GT28_MurG | 5.58e-08 | 34 | 249 | 151 | 347 | undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase. MurG (EC 2.4.1.227) is an N-acetylglucosaminyltransferase, the last enzyme involved in the intracellular phase of peptidoglycan biosynthesis. It transfers N-acetyl-D-glucosamine (GlcNAc) from UDP-GlcNAc to the C4 hydroxyl of a lipid-linked N-acetylmuramoyl pentapeptide (NAM). The resulting disaccharide is then transported across the cell membrane, where it is polymerized into NAG-NAM cell-wall repeat structure. MurG belongs to the GT-B structural superfamily of glycoslytransferases, which have characteristic N- and C-terminal domains, each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.53e-49 | 18 | 283 | 227 | 502 | |
| 1.56e-49 | 18 | 284 | 228 | 504 | |
| 2.01e-48 | 18 | 283 | 225 | 500 | |
| 2.18e-48 | 18 | 283 | 227 | 502 | |
| 2.18e-48 | 18 | 283 | 227 | 502 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.17e-13 | 41 | 231 | 19 | 186 | Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_B Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_C Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_D Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens] |
|
| 8.60e-12 | 50 | 228 | 7 | 162 | Chain A, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_B Chain B, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_C Chain C, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_D Chain D, UDP-glucuronosyltransferase 2B15 [Homo sapiens] |
|
| 3.25e-11 | 50 | 231 | 6 | 164 | Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens],2O6L_B Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens] |
|
| 1.85e-09 | 135 | 198 | 279 | 340 | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_B Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_C Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_D Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora] |
|
| 1.86e-09 | 135 | 198 | 282 | 343 | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_B Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_C Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_D Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.81e-18 | 16 | 303 | 248 | 505 | UDP-glucuronosyltransferase 1A7 OS=Homo sapiens OX=9606 GN=UGT1A7 PE=1 SV=2 |
|
| 5.81e-18 | 16 | 303 | 248 | 505 | UDP-glucuronosyltransferase 1A9 OS=Homo sapiens OX=9606 GN=UGT1A9 PE=1 SV=1 |
|
| 5.81e-18 | 16 | 303 | 248 | 505 | UDP-glucuronosyltransferase 1A8 OS=Homo sapiens OX=9606 GN=UGT1A8 PE=1 SV=1 |
|
| 5.81e-18 | 16 | 303 | 248 | 505 | UDP-glucuronosyltransferase 1A10 OS=Homo sapiens OX=9606 GN=UGT1A10 PE=1 SV=1 |
|
| 7.83e-18 | 21 | 257 | 259 | 474 | UDP-glucuronosyltransferase 2B14 OS=Oryctolagus cuniculus OX=9986 GN=UGT2B14 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
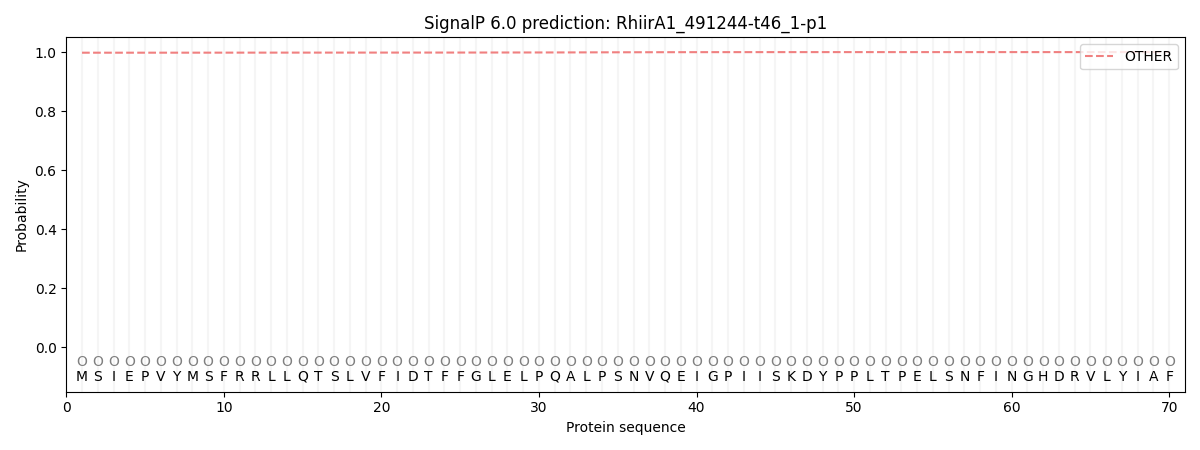
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998139 | 0.001861 |