You are browsing environment: FUNGIDB
CAZyme Information: RhiirA1_442609-t46_1-p1
You are here: Home > Sequence: RhiirA1_442609-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
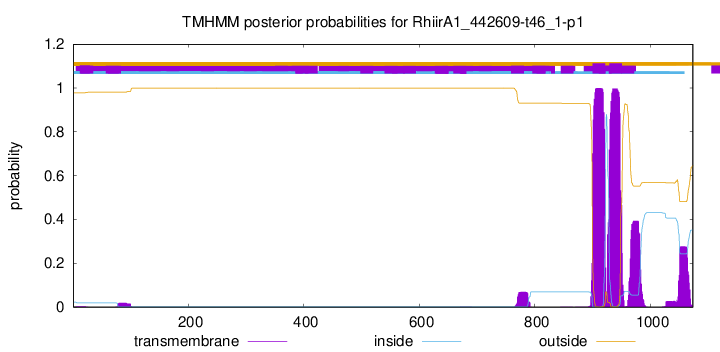
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | RhiirA1_442609-t46_1-p1 | |||||||||||
| CAZy Family | PL38 | |||||||||||
| CAZyme Description | NAD(P)-binding protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178298 | PLN02695 | 7.97e-92 | 3 | 354 | 19 | 336 | GDP-D-mannose-3',5'-epimerase |
| 187581 | GME-like_SDR_e | 1.70e-87 | 8 | 355 | 3 | 320 | Arabidopsis thaliana GDP-mannose-3',5'-epimerase (GME)-like, extended (e) SDRs. This subgroup of NDP-sugar epimerase/dehydratases are extended SDRs; they have the characteristic active site tetrad, and an NAD-binding motif: TGXXGXX[AG], which is a close match to the canonical NAD-binding motif. Members include Arabidopsis thaliana GDP-mannose-3',5'-epimerase (GME) which catalyzes the epimerization of two positions of GDP-alpha-D-mannose to form GDP-beta-L-galactose. Extended SDRs are distinct from classical SDRs. In addition to the Rossmann fold (alpha/beta folding pattern with a central beta-sheet) core region typical of all SDRs, extended SDRs have a less conserved C-terminal extension of approximately 100 amino acids. Extended SDRs are a diverse collection of proteins, and include isomerases, epimerases, oxidoreductases, and lyases; they typically have a TGXXGXXG cofactor binding motif. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold, an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Sequence identity between different SDR enzymes is typically in the 15-30% range; they catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase numbering). In addition to the Tyr and Lys, there is often an upstream Ser and/or an Asn, contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Atypical SDRs generally lack the catalytic residues characteristic of the SDRs, and their glycine-rich NAD(P)-binding motif is often different from the forms normally seen in classical or extended SDRs. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. |
| 223528 | WcaG | 1.90e-47 | 8 | 353 | 3 | 314 | Nucleoside-diphosphate-sugar epimerase [Cell wall/membrane/envelope biogenesis]. |
| 187566 | UDP_AE_SDR_e | 3.28e-40 | 8 | 347 | 2 | 304 | UDP-N-acetylglucosamine 4-epimerase, extended (e) SDRs. This subgroup contains UDP-N-acetylglucosamine 4-epimerase of Pseudomonas aeruginosa, WbpP, an extended SDR, that catalyzes the NAD+ dependent conversion of UDP-GlcNAc and UDPGalNA to UDP-Glc and UDP-Gal. This subgroup has the characteristic active site tetrad and NAD-binding motif of the extended SDRs. Extended SDRs are distinct from classical SDRs. In addition to the Rossmann fold (alpha/beta folding pattern with a central beta-sheet) core region typical of all SDRs, extended SDRs have a less conserved C-terminal extension of approximately 100 amino acids. Extended SDRs are a diverse collection of proteins, and include isomerases, epimerases, oxidoreductases, and lyases; they typically have a TGXXGXXG cofactor binding motif. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold, an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Sequence identity between different SDR enzymes is typically in the 15-30% range; they catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase numbering). In addition to the Tyr and Lys, there is often an upstream Ser and/or an Asn, contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Atypical SDRs generally lack the catalytic residues characteristic of the SDRs, and their glycine-rich NAD(P)-binding motif is often different from the forms normally seen in classical or extended SDRs. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. |
| 396097 | Epimerase | 2.89e-38 | 8 | 277 | 1 | 238 | NAD dependent epimerase/dehydratase family. This family of proteins utilize NAD as a cofactor. The proteins in this family use nucleotide-sugar substrates for a variety of chemical reactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.88e-279 | 7 | 1033 | 18 | 1011 | |
| 4.77e-275 | 6 | 1029 | 17 | 1007 | |
| 3.57e-261 | 6 | 1029 | 18 | 1011 | |
| 5.94e-223 | 7 | 989 | 13 | 924 | |
| 6.66e-35 | 646 | 901 | 25 | 272 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.58e-67 | 3 | 362 | 27 | 353 | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), with gdp-alpha-d-mannose and gdp-beta-l-galactose bound in the active site. [Arabidopsis thaliana],2C59_B gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), with gdp-alpha-d-mannose and gdp-beta-l-galactose bound in the active site. [Arabidopsis thaliana] |
|
| 1.18e-66 | 3 | 362 | 27 | 353 | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana),k178r, with gdp-beta-l-gulose and gdp-4-keto-beta-l-gulose bound in active site. [Arabidopsis thaliana],2C54_B gdp-mannose-3', 5' -epimerase (arabidopsis thaliana),k178r, with gdp-beta-l-gulose and gdp-4-keto-beta-l-gulose bound in active site. [Arabidopsis thaliana] |
|
| 1.62e-66 | 3 | 362 | 27 | 353 | GDP-mannose-3', 5' -epimerase (Arabidopsis thaliana),Y174F, with GDP-beta-L-galactose bound in the active site [Arabidopsis thaliana],2C5A_B GDP-mannose-3', 5' -epimerase (Arabidopsis thaliana),Y174F, with GDP-beta-L-galactose bound in the active site [Arabidopsis thaliana] |
|
| 3.04e-66 | 3 | 362 | 27 | 353 | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), k217a, with gdp-alpha-d-mannose bound in the active site. [Arabidopsis thaliana],2C5E_B gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), k217a, with gdp-alpha-d-mannose bound in the active site. [Arabidopsis thaliana] |
|
| 6.30e-21 | 8 | 352 | 6 | 312 | Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) from Hyperthermophilic Eubacterium Thermotoga maritima [Thermotoga maritima MSB8],4ZRM_B Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) from Hyperthermophilic Eubacterium Thermotoga maritima [Thermotoga maritima MSB8],4ZRN_A Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) with UDP-glucose from Hyperthermophilic Eubacterium Thermotoga Maritima [Thermotoga maritima MSB8],4ZRN_B Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) with UDP-glucose from Hyperthermophilic Eubacterium Thermotoga Maritima [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.44e-68 | 3 | 360 | 27 | 350 | GDP-mannose 3,5-epimerase 1 OS=Oryza sativa subsp. indica OX=39946 GN=OsI_032456 PE=2 SV=1 |
|
| 1.98e-68 | 3 | 360 | 27 | 350 | GDP-mannose 3,5-epimerase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=GME-1 PE=1 SV=1 |
|
| 2.23e-66 | 3 | 362 | 25 | 351 | GDP-mannose 3,5-epimerase OS=Arabidopsis thaliana OX=3702 GN=At5g28840 PE=1 SV=1 |
|
| 4.84e-66 | 4 | 355 | 21 | 338 | GDP-mannose 3,5-epimerase 2 OS=Oryza sativa subsp. japonica OX=39947 GN=GME-2 PE=2 SV=2 |
|
| 8.22e-22 | 8 | 343 | 3 | 297 | GDP-L-colitose synthase OS=Yersinia pseudotuberculosis OX=633 GN=colC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
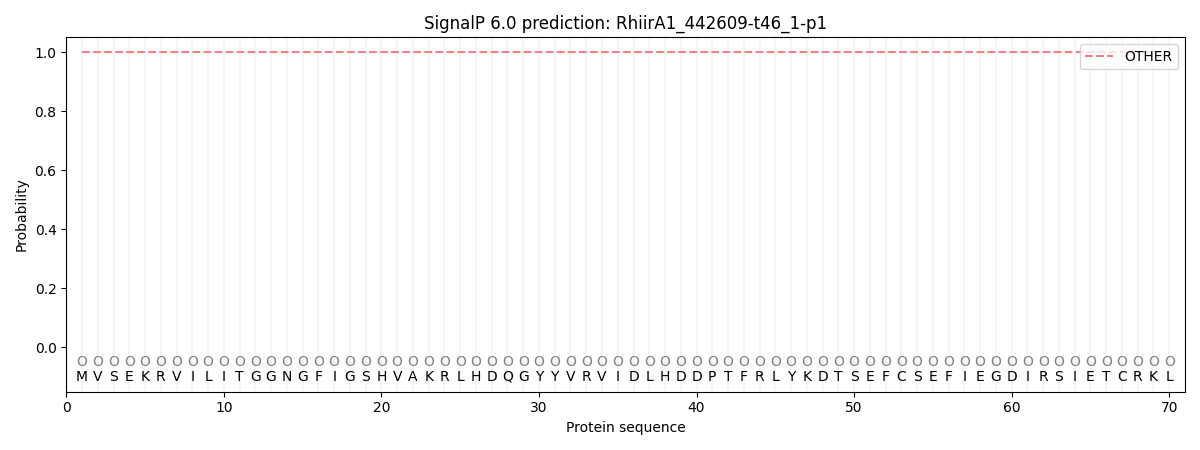
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000076 | 0.000000 |