You are browsing environment: FUNGIDB
CAZyme Information: RVD90391.1
You are here: Home > Sequence: RVD90391.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD90391.1 | |||||||||||
| CAZy Family | GT92 | |||||||||||
| CAZyme Description | CBM1 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A437AGY4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH131 | 19 | 261 | 6.8e-103 | 0.996078431372549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408085 | GH131_N | 8.90e-82 | 19 | 252 | 1 | 248 | Glycoside hydrolase 131 catalytic N-terminal domain. This is the N-terminal domain found in glycoside hydrolase family 131 (GH131A) protein observed in Coprinopsis cinerea. GH131A exhibits bifunctional exo-beta-1,3-/-1,6- and endo-beta-1,4 activity toward beta-glucan. This domain is catalytic in nature though the catalytic mechanism of C. cinerea GH131A is different from that of typical glycosidases that use a pair of carboxylic acid residues as the catalytic residues. In the case of GH131A, Glu98 and His218 may form a catalytic dyad and Glu98 may activate His218 during catalysis. |
| 197593 | fCBD | 2.19e-13 | 313 | 345 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 3.16e-13 | 313 | 341 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.25e-203 | 1 | 345 | 1 | 369 | |
| 5.00e-136 | 6 | 344 | 6 | 337 | |
| 9.67e-136 | 9 | 344 | 11 | 346 | |
| 1.89e-133 | 4 | 344 | 6 | 347 | |
| 1.40e-126 | 3 | 260 | 4 | 259 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.82e-125 | 17 | 264 | 1 | 246 | Chain A, Beta-glucanase [Podospora anserina],4LE3_B Chain B, Beta-glucanase [Podospora anserina],4LE3_C Chain C, Beta-glucanase [Podospora anserina],4LE3_D Chain D, Beta-glucanase [Podospora anserina],4LE4_A Chain A, Beta-glucanase [Podospora anserina],4LE4_B Chain B, Beta-glucanase [Podospora anserina],4LE4_C Chain C, Beta-glucanase [Podospora anserina],4LE4_D Chain D, Beta-glucanase [Podospora anserina] |
|
| 2.90e-91 | 17 | 261 | 1 | 234 | Chain A, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_B Chain B, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_C Chain C, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_D Chain D, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130] |
|
| 3.18e-12 | 311 | 345 | 2 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 1.13e-11 | 314 | 345 | 5 | 36 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 1.55e-11 | 315 | 345 | 6 | 36 | Chain A, CELLOBIOHYDROLASE I [Trichoderma reesei],1AZH_A Chain A, CELLOBIOHYDROLASE I [Trichoderma reesei],5X3C_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.37e-11 | 311 | 345 | 19 | 53 | Manganese dependent endoglucanase Eg5A OS=Phanerodontia chrysosporium OX=2822231 GN=Eg5A PE=1 SV=2 |
|
| 7.28e-11 | 309 | 345 | 17 | 54 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=cbhC PE=3 SV=1 |
|
| 9.80e-11 | 312 | 345 | 23 | 56 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.18.61.1 PE=1 SV=1 |
|
| 2.78e-10 | 311 | 345 | 23 | 57 | Endoglucanase EG-II OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=egl2 PE=1 SV=1 |
|
| 2.78e-10 | 311 | 345 | 23 | 57 | Endoglucanase EG-II OS=Hypocrea jecorina OX=51453 GN=egl2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
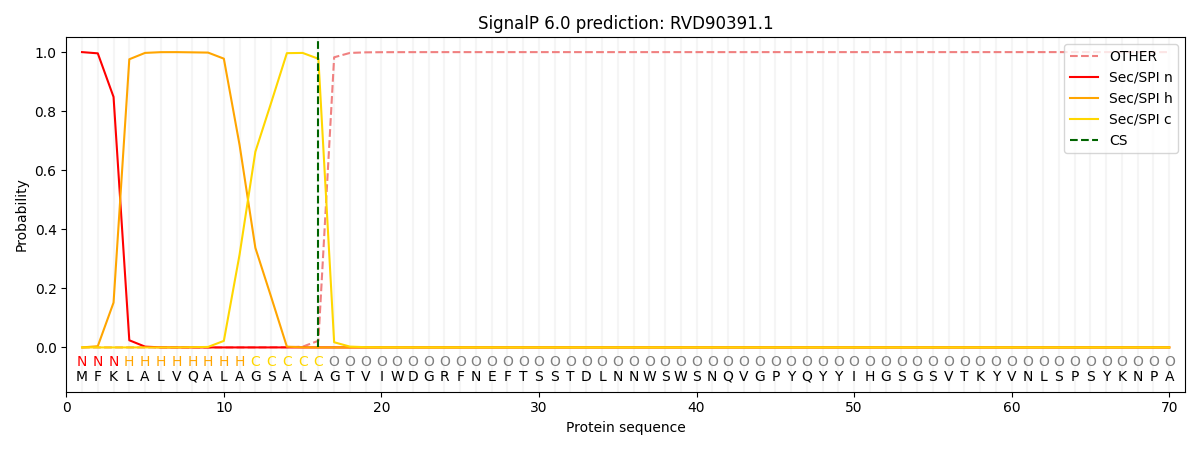
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000273 | 0.999690 | CS pos: 16-17. Pr: 0.9774 |
