You are browsing environment: FUNGIDB
CAZyme Information: RVD85360.1
You are here: Home > Sequence: RVD85360.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD85360.1 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | Cutinase [Source:UniProtKB/TrEMBL;Acc:A0A437A2Q0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 46 | 219 | 3.7e-40 | 0.9841269841269841 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 3.73e-55 | 46 | 220 | 2 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.08e-145 | 1 | 226 | 1 | 226 | |
| 1.59e-87 | 1 | 220 | 1 | 224 | |
| 6.70e-87 | 1 | 205 | 1 | 197 | |
| 9.69e-79 | 5 | 221 | 6 | 216 | |
| 9.67e-77 | 5 | 223 | 6 | 217 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.17e-57 | 25 | 220 | 3 | 198 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 6.36e-55 | 44 | 222 | 29 | 209 | Chain A, CUTINASE [Fusarium vanettenii],1CUW_B Chain B, CUTINASE [Fusarium vanettenii] |
|
| 1.81e-54 | 44 | 222 | 29 | 209 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 5.57e-54 | 44 | 222 | 15 | 194 | Humicola insolens cutinase [Humicola insolens],4OYY_B Humicola insolens cutinase [Humicola insolens],4OYY_C Humicola insolens cutinase [Humicola insolens],4OYY_D Humicola insolens cutinase [Humicola insolens],4OYY_E Humicola insolens cutinase [Humicola insolens],4OYY_F Humicola insolens cutinase [Humicola insolens],4OYY_G Humicola insolens cutinase [Humicola insolens],4OYY_H Humicola insolens cutinase [Humicola insolens],4OYY_I Humicola insolens cutinase [Humicola insolens],4OYY_J Humicola insolens cutinase [Humicola insolens],4OYY_K Humicola insolens cutinase [Humicola insolens],4OYY_L Humicola insolens cutinase [Humicola insolens] |
|
| 6.70e-54 | 44 | 222 | 15 | 195 | The 1.15 angstrom refined structure of fusarium solani pisi cutinase [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.24e-66 | 1 | 220 | 1 | 214 | Cutinase CUT2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=CUT2 PE=1 SV=1 |
|
| 1.90e-63 | 1 | 223 | 3 | 224 | Probable cutinase 1 OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=AFLA_039350 PE=3 SV=1 |
|
| 7.65e-63 | 1 | 223 | 3 | 224 | Probable cutinase 3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=AO090011000113 PE=3 SV=1 |
|
| 1.54e-62 | 1 | 220 | 3 | 213 | Cutinase 1 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cut1 PE=2 SV=1 |
|
| 8.85e-61 | 5 | 220 | 7 | 218 | Probable cutinase 5 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=ATEG_03640 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
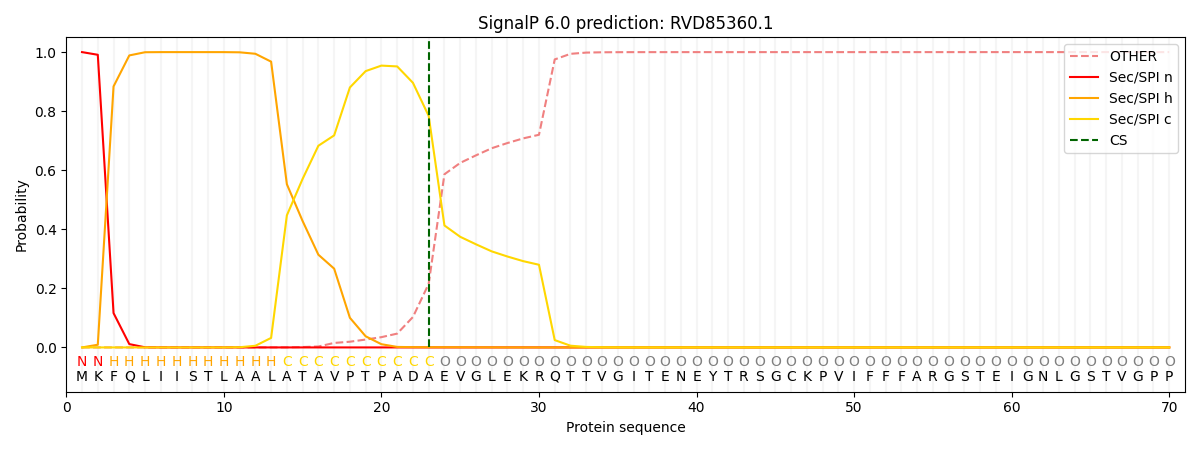
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000244 | 0.999715 | CS pos: 23-24. Pr: 0.7839 |
