You are browsing environment: FUNGIDB
CAZyme Information: RVD83828.1
You are here: Home > Sequence: RVD83828.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD83828.1 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 20 | 456 | 1.6e-183 | 0.9932279909706546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227807 | XynC | 1.41e-23 | 7 | 428 | 20 | 430 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| 405300 | Glyco_hydr_30_2 | 1.28e-16 | 22 | 279 | 3 | 285 | O-Glycosyl hydrolase family 30. |
| 395595 | CBM_1 | 9.27e-13 | 494 | 522 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 8.51e-12 | 494 | 526 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 407314 | Glyco_hydro_30C | 4.06e-09 | 370 | 456 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 526 | 1 | 533 | |
| 5.58e-240 | 11 | 458 | 11 | 458 | |
| 2.03e-236 | 11 | 458 | 15 | 462 | |
| 3.22e-234 | 6 | 458 | 5 | 457 | |
| 3.87e-233 | 1 | 458 | 1 | 458 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.66e-148 | 22 | 455 | 24 | 450 | Chain A, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
|
| 1.69e-116 | 19 | 458 | 1 | 447 | Chain A, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 3.41e-115 | 19 | 458 | 8 | 454 | Chain AAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 1.12e-101 | 14 | 411 | 17 | 421 | Chain A, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
|
| 8.61e-101 | 15 | 411 | 87 | 492 | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.38e-116 | 19 | 458 | 25 | 471 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
|
| 6.23e-13 | 23 | 404 | 87 | 484 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
|
| 1.80e-11 | 24 | 401 | 88 | 479 | Putative glucosylceramidase 2 OS=Caenorhabditis elegans OX=6239 GN=gba-2 PE=3 SV=2 |
|
| 1.29e-10 | 16 | 401 | 81 | 483 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
|
| 4.13e-10 | 491 | 526 | 506 | 541 | Probable 1,4-beta-D-glucan cellobiohydrolase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=cbhB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
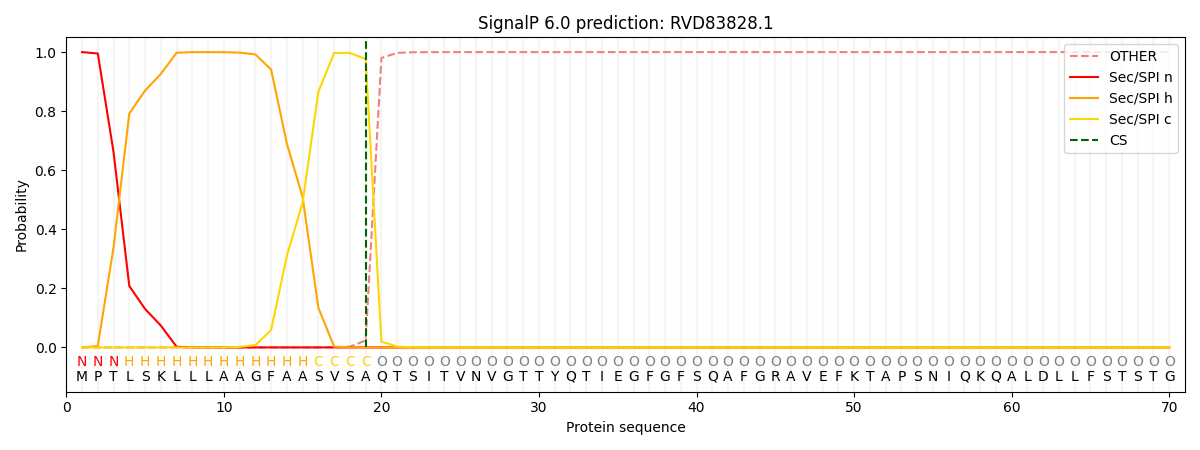
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000266 | 0.999718 | CS pos: 19-20. Pr: 0.9765 |
