You are browsing environment: FUNGIDB
CAZyme Information: RVD83421.1
You are here: Home > Sequence: RVD83421.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD83421.1 | |||||||||||
| CAZy Family | CE9 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase [Source:UniProtKB/TrEMBL;Acc:A0A436ZWQ4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 44 | 220 | 4.7e-73 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 7.26e-104 | 44 | 219 | 1 | 175 | Glycosyl hydrolases family 11. |
| 197593 | fCBD | 4.45e-12 | 256 | 289 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 1.20e-11 | 257 | 285 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.69e-166 | 1 | 289 | 1 | 292 | |
| 1.35e-122 | 1 | 289 | 1 | 282 | |
| 1.05e-118 | 1 | 289 | 1 | 278 | |
| 1.05e-118 | 1 | 289 | 1 | 278 | |
| 1.66e-116 | 1 | 289 | 1 | 293 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.59e-100 | 34 | 222 | 4 | 192 | High resolution structure of GH11 xylanase from Nectria haematococca [Fusarium vanettenii 77-13-4] |
|
| 2.70e-95 | 36 | 222 | 7 | 192 | Crystal Structure of a Xylanase at 1.56 Angstroem resolution [Fusarium oxysporum],5JRN_A Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution [Fusarium oxysporum f. sp. vasinfectum 25433] |
|
| 1.37e-90 | 36 | 222 | 3 | 190 | ENDO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.0 [Thermomyces lanuginosus] |
|
| 5.21e-88 | 36 | 222 | 3 | 190 | Chain A, Protein (endo-1,4-beta-xylanase) [Paecilomyces variotii] |
|
| 9.16e-88 | 40 | 222 | 7 | 189 | HIGH-RESOLUTION STRUCTURES OF XYLANASES FROM B. CIRCULANS AND T. HARZIANUM IDENTIFY A NEW FOLDING PATTERN AND IMPLICATIONS FOR THE ATOMIC BASIS OF THE CATALYSIS [Trichoderma harzianum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.30e-104 | 1 | 222 | 1 | 229 | Endo-1,4-beta-xylanase 4 OS=Magnaporthe grisea OX=148305 GN=XYL4 PE=3 SV=2 |
|
| 3.30e-104 | 1 | 222 | 1 | 229 | Endo-1,4-beta-xylanase 4 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL4 PE=3 SV=1 |
|
| 1.34e-103 | 1 | 222 | 1 | 218 | Endo-1,4-beta-xylanase I OS=Cochliobolus carbonum OX=5017 GN=XYL1 PE=1 SV=1 |
|
| 6.97e-103 | 1 | 289 | 1 | 290 | Endo-1,4-beta-xylanase B OS=Phanerodontia chrysosporium OX=2822231 GN=xynB PE=1 SV=1 |
|
| 1.71e-97 | 1 | 222 | 1 | 222 | Effector Vd424Y OS=Verticillium dahliae (strain VdLs.17 / ATCC MYA-4575 / FGSC 10137) OX=498257 GN=VDAG_05042 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
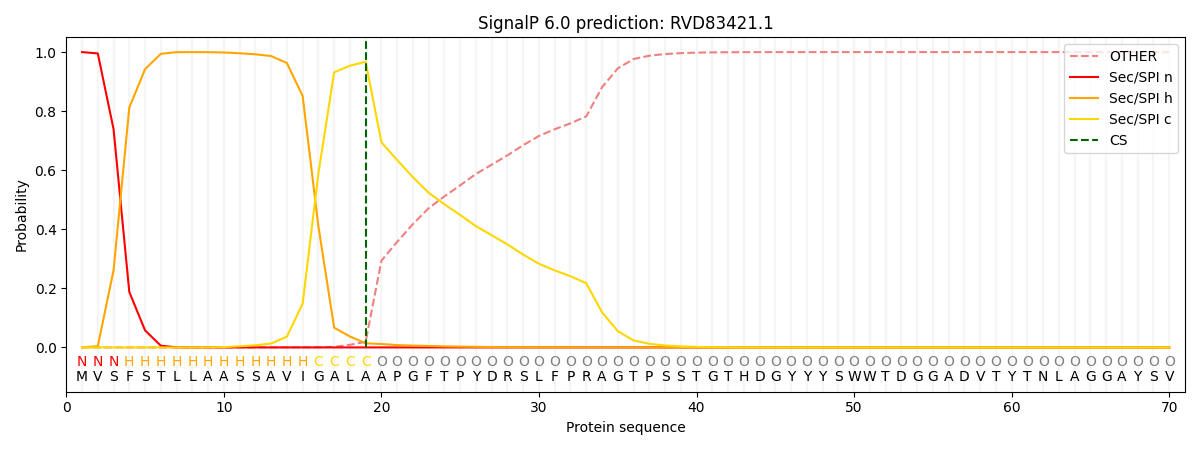
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000550 | 0.999445 | CS pos: 19-20. Pr: 0.9673 |
