You are browsing environment: FUNGIDB
CAZyme Information: RVD83167.1
You are here: Home > Sequence: RVD83167.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD83167.1 | |||||||||||
| CAZy Family | CE18 | |||||||||||
| CAZyme Description | Beta-xylanase [Source:UniProtKB/TrEMBL;Acc:A0A436ZW22] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 64 | 374 | 4.5e-89 | 0.9603960396039604 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 1.11e-121 | 67 | 374 | 12 | 310 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 3.50e-100 | 99 | 372 | 1 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 1.02e-74 | 89 | 374 | 57 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.67e-280 | 1 | 391 | 1 | 392 | |
| 3.83e-109 | 65 | 376 | 37 | 343 | |
| 3.08e-104 | 65 | 376 | 48 | 349 | |
| 8.75e-104 | 52 | 376 | 35 | 349 | |
| 1.20e-103 | 65 | 376 | 45 | 346 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.51e-93 | 57 | 376 | 6 | 312 | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 2.43e-92 | 57 | 376 | 6 | 312 | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 3.03e-89 | 65 | 372 | 13 | 313 | Crystal structure of GH10 family xylanase XynAF1 from Aspergillus fumigatus Z5 [Aspergillus fumigatus Z5],6JDT_B Crystal structure of GH10 family xylanase XynAF1 from Aspergillus fumigatus Z5 [Aspergillus fumigatus Z5],6JDY_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 120 minutes [Aspergillus fumigatus Z5],6JDY_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 120 minutes [Aspergillus fumigatus Z5],6JDZ_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 20 minutes [Aspergillus fumigatus Z5],6JDZ_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 20 minutes [Aspergillus fumigatus Z5],6JE0_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 30 minutes [Aspergillus fumigatus Z5],6JE0_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 30 minutes [Aspergillus fumigatus Z5],6JE1_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 40 minutes [Aspergillus fumigatus Z5],6JE1_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 40 minutes [Aspergillus fumigatus Z5],6JE2_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 80 minutes [Aspergillus fumigatus Z5],6JE2_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 80 minutes [Aspergillus fumigatus Z5] |
|
| 3.12e-88 | 65 | 372 | 35 | 335 | GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8M_B GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8N_A GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872],6Q8N_B GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872] |
|
| 5.95e-79 | 74 | 377 | 48 | 345 | Chain A, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.80e-90 | 64 | 374 | 28 | 329 | Endo-1,4-beta-xylanase OS=Agaricus bisporus OX=5341 GN=xlnA PE=2 SV=1 |
|
| 1.23e-89 | 57 | 376 | 44 | 350 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
|
| 2.07e-86 | 66 | 372 | 95 | 393 | Endo-1,4-beta-xylanase C OS=Phanerodontia chrysosporium OX=2822231 GN=xynC PE=1 SV=1 |
|
| 1.07e-85 | 66 | 372 | 105 | 402 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
|
| 4.01e-79 | 66 | 372 | 32 | 330 | Endo-1,4-beta-xylanase D OS=Talaromyces funiculosus OX=28572 GN=xynD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
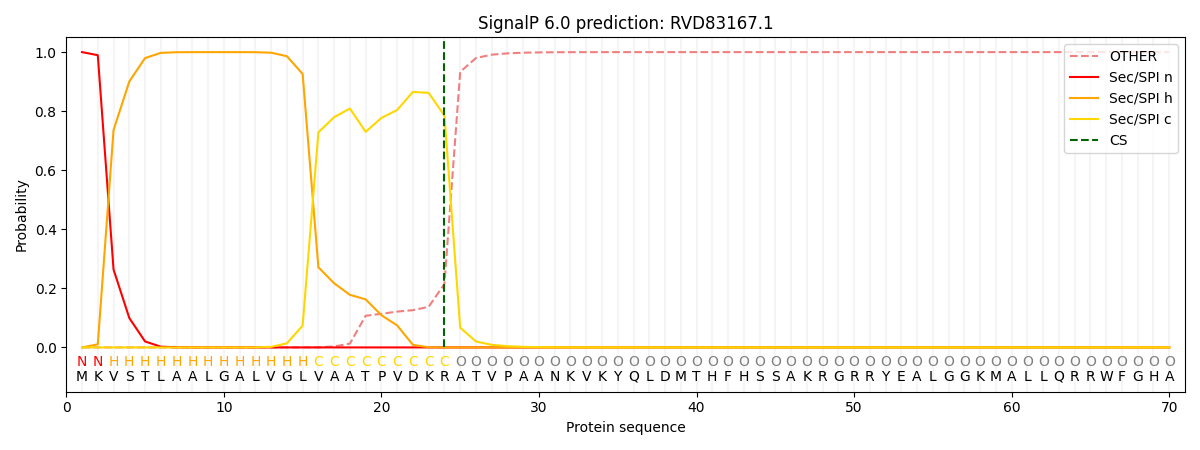
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000497 | 0.999471 | CS pos: 24-25. Pr: 0.7849 |
