You are browsing environment: FUNGIDB
CAZyme Information: RVD82459.1
You are here: Home > Sequence: RVD82459.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
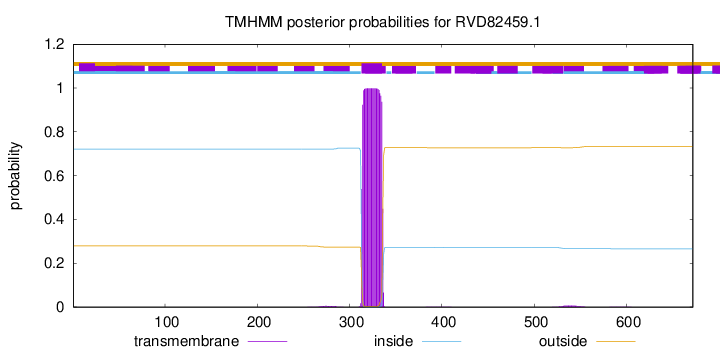
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD82459.1 | |||||||||||
| CAZy Family | CBM67|GH78 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 420 | 660 | 7.2e-29 | 0.8971061093247589 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.40e-33 | 412 | 657 | 68 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 2.18e-07 | 471 | 660 | 76 | 304 | Glycosyl hydrolases family 17. |
| 236669 | PRK10263 | 7.42e-05 | 4 | 186 | 360 | 522 | DNA translocase FtsK; Provisional |
| 312941 | Med15 | 5.17e-04 | 5 | 187 | 157 | 355 | ARC105 or Med15 subunit of Mediator complex non-fungal. The approx. 70 residue Med15 domain of the ARC-Mediator co-activator is a three-helix bundle with marked similarity to the KIX domain. The sterol regulatory element binding protein (SREBP) family of transcription activators use the ARC105 subunit to activate target genes in the regulation of cholesterol and fatty acid homeostasis. In addition, Med15 is a critical transducer of gene activation signals that control early metazoan development. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 664 | 1 | 664 | |
| 3.49e-109 | 368 | 663 | 343 | 646 | |
| 1.56e-108 | 364 | 663 | 361 | 667 | |
| 4.41e-107 | 364 | 663 | 339 | 645 | |
| 4.67e-107 | 364 | 663 | 401 | 706 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.16e-31 | 382 | 664 | 35 | 293 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 2.01e-30 | 382 | 664 | 35 | 293 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.16e-100 | 364 | 663 | 361 | 674 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgC PE=3 SV=1 |
|
| 1.16e-100 | 364 | 663 | 361 | 674 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgC PE=3 SV=2 |
|
| 1.70e-98 | 368 | 663 | 335 | 644 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgC PE=3 SV=2 |
|
| 1.78e-98 | 363 | 663 | 359 | 673 | Putative glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=btgC PE=3 SV=2 |
|
| 1.65e-96 | 364 | 663 | 325 | 638 | Glucan endo-1,3-beta-glucosidase btgC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=btgC PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000047 | 0.000000 |