You are browsing environment: FUNGIDB
CAZyme Information: RVD80878.1
You are here: Home > Sequence: RVD80878.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arthrobotrys flagrans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Orbiliomycetes; ; Orbiliaceae; Arthrobotrys; Arthrobotrys flagrans | |||||||||||
| CAZyme ID | RVD80878.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Pectinesterase [Source:UniProtKB/TrEMBL;Acc:A0A436ZPQ6] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.11:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 44 | 318 | 1e-72 | 0.9131944444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395871 | Pectinesterase | 4.48e-50 | 41 | 312 | 1 | 266 | Pectinesterase. |
| 178051 | PLN02432 | 9.62e-42 | 42 | 334 | 13 | 287 | putative pectinesterase |
| 215357 | PLN02665 | 1.65e-40 | 54 | 324 | 80 | 341 | pectinesterase family protein |
| 215170 | PLN02301 | 2.95e-40 | 40 | 312 | 236 | 502 | pectinesterase/pectinesterase inhibitor |
| 178106 | PLN02488 | 2.06e-38 | 13 | 313 | 167 | 464 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.49e-220 | 1 | 348 | 1 | 349 | |
| 8.00e-119 | 10 | 348 | 10 | 338 | |
| 6.77e-117 | 27 | 348 | 1302 | 1619 | |
| 1.76e-101 | 1 | 348 | 1 | 344 | |
| 4.99e-101 | 1 | 348 | 1 | 344 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.15e-57 | 53 | 347 | 18 | 299 | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
|
| 5.85e-57 | 53 | 347 | 18 | 299 | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
|
| 8.28e-33 | 54 | 312 | 19 | 273 | Pectin methylesterase from Carrot [Daucus carota] |
|
| 2.16e-29 | 45 | 312 | 10 | 269 | Chain A, Pectinesterase 1 [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.70e-76 | 42 | 348 | 28 | 330 | Pectinesterase OS=Aspergillus niger OX=5061 GN=pme1 PE=1 SV=1 |
|
| 1.71e-73 | 13 | 347 | 1 | 330 | Pectinesterase OS=Aspergillus aculeatus OX=5053 GN=pme1 PE=2 SV=1 |
|
| 1.07e-63 | 42 | 347 | 36 | 324 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
|
| 2.12e-63 | 54 | 347 | 44 | 324 | Probable pectinesterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pmeA PE=3 SV=1 |
|
| 3.00e-63 | 42 | 347 | 36 | 324 | Probable pectinesterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
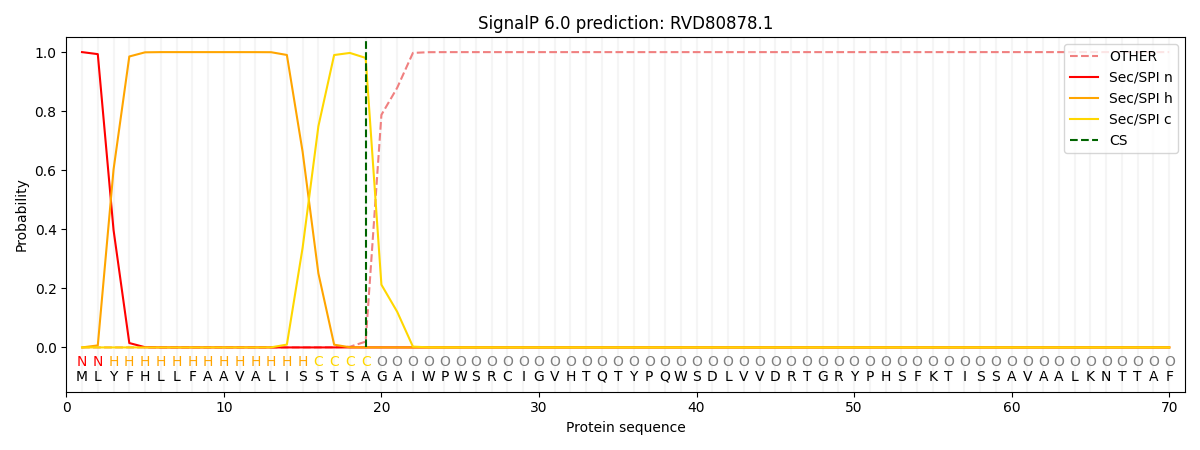
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000253 | 0.999706 | CS pos: 19-20. Pr: 0.9803 |
