You are browsing environment: FUNGIDB
CAZyme Information: RQM13996.1
You are here: Home > Sequence: RQM13996.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Peronospora effusa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Peronospora; Peronospora effusa | |||||||||||
| CAZyme ID | RQM13996.1 | |||||||||||
| CAZy Family | GH5|CBM43 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 11 | 260 | 1.8e-61 | 0.9306122448979591 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 411345 | gliding_GltJ | 5.31e-05 | 247 | 331 | 424 | 505 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 411100 | internalin_K | 1.08e-04 | 247 | 292 | 513 | 558 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
| 236776 | PRK10856 | 3.59e-04 | 247 | 316 | 167 | 235 | cytoskeleton protein RodZ. |
| 236776 | PRK10856 | 6.82e-04 | 239 | 315 | 177 | 252 | cytoskeleton protein RodZ. |
| 405911 | TALPID3 | 0.001 | 247 | 325 | 944 | 1031 | Hedgehog signalling target. TALPID3 is a family of eukaryotic proteins that are targets for Hedgehog signalling. Mutations in this gene noticed first in chickens lead to multiple abnormalities of development. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.24e-180 | 1 | 331 | 1 | 363 | |
| 4.11e-130 | 1 | 216 | 1 | 216 | |
| 7.75e-104 | 1 | 219 | 1 | 218 | |
| 3.13e-103 | 1 | 219 | 1 | 218 | |
| 4.44e-103 | 1 | 219 | 1 | 218 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.21e-15 | 22 | 191 | 1 | 170 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
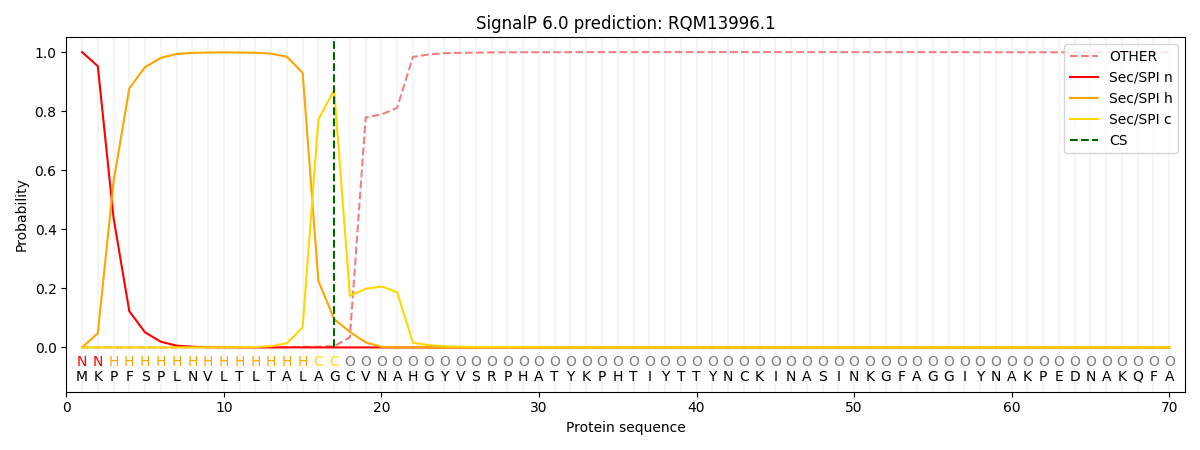
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000217 | 0.999782 | CS pos: 17-18. Pr: 0.8717 |
