You are browsing environment: FUNGIDB
CAZyme Information: RQM13721.1
You are here: Home > Sequence: RQM13721.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Peronospora effusa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Peronospora; Peronospora effusa | |||||||||||
| CAZyme ID | RQM13721.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 58 | 534 | 6.4e-87 | 0.9385474860335196 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 1.04e-70 | 59 | 542 | 34 | 526 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 177843 | PLN02191 | 2.39e-63 | 59 | 542 | 56 | 549 | L-ascorbate oxidase |
| 215324 | PLN02604 | 5.75e-58 | 64 | 539 | 62 | 546 | oxidoreductase |
| 225043 | SufI | 4.79e-56 | 38 | 541 | 46 | 450 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| 274556 | laccase | 1.33e-45 | 64 | 538 | 41 | 518 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 569 | 1 | 569 | |
| 0.0 | 7 | 569 | 5 | 571 | |
| 6.10e-190 | 13 | 568 | 13 | 551 | |
| 3.45e-177 | 11 | 568 | 4 | 543 | |
| 2.58e-79 | 39 | 561 | 55 | 553 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.32e-56 | 3 | 554 | 2 | 502 | Chain A, Laccase [Rigidoporus microporus] |
|
| 2.61e-56 | 64 | 539 | 41 | 465 | Crystal Structure of Laccase from Cerrena sp. RSD1 [Cerrena],5Z1X_B Crystal Structure of Laccase from Cerrena sp. RSD1 [Cerrena],5Z22_A Crystal Structure of Laccase from Cerrena sp. RSD1 [Cerrena] |
|
| 9.89e-56 | 64 | 541 | 105 | 544 | Structure of the L499M mutant of the laccase from B.aclada [Botrytis aclada] |
|
| 1.60e-55 | 65 | 539 | 42 | 468 | Chain A, LACCASE 1 [Coprinopsis cinerea] |
|
| 1.63e-55 | 65 | 539 | 42 | 468 | Chain A, Laccase [Coprinopsis cinerea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.57e-64 | 15 | 541 | 52 | 523 | Laccase-1 OS=Botryotinia fuckeliana OX=40559 GN=lcc1 PE=2 SV=3 |
|
| 3.55e-62 | 21 | 568 | 20 | 518 | Iron transport multicopper oxidase fetC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=fetC PE=2 SV=1 |
|
| 3.09e-58 | 19 | 566 | 18 | 518 | Iron transport multicopper oxidase fetC OS=Epichloe festucae (strain E2368) OX=696363 GN=fetC PE=2 SV=1 |
|
| 7.47e-57 | 59 | 541 | 101 | 545 | Laccase-2 OS=Botryotinia fuckeliana OX=40559 GN=lcc2 PE=2 SV=1 |
|
| 1.53e-56 | 64 | 543 | 75 | 502 | Laccase-2 OS=Pleurotus ostreatus OX=5322 GN=POX2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
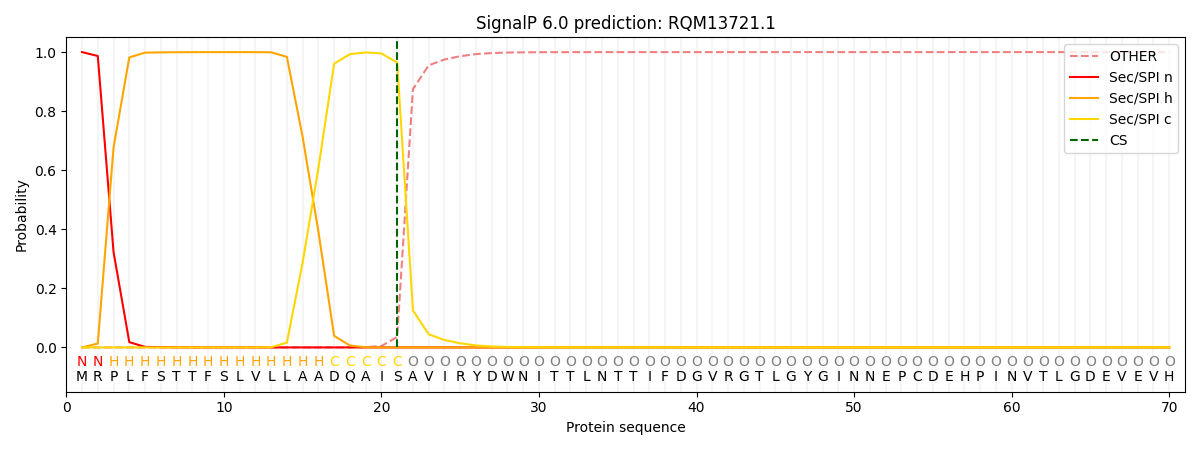
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000687 | 0.999290 | CS pos: 21-22. Pr: 0.9645 |
