You are browsing environment: FUNGIDB
CAZyme Information: RQM12451.1
You are here: Home > Sequence: RQM12451.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Peronospora effusa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Peronospora; Peronospora effusa | |||||||||||
| CAZyme ID | RQM12451.1 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 488 | 739 | 1.1e-66 | 0.9224489795918367 |
| AA17 | 17 | 262 | 1e-60 | 0.9142857142857143 |
| AA17 | 297 | 477 | 1.1e-44 | 0.6693877551020408 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 183854 | PRK13042 | 0.001 | 696 | 771 | 25 | 99 | superantigen-like protein SSL4; Reviewed. |
| 236090 | PRK07764 | 0.001 | 693 | 762 | 431 | 500 | DNA polymerase III subunits gamma and tau; Validated |
| 237871 | PRK14965 | 0.002 | 721 | 764 | 390 | 433 | DNA polymerase III subunits gamma and tau; Provisional |
| 411345 | gliding_GltJ | 0.002 | 693 | 764 | 405 | 462 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 235899 | PRK06975 | 0.003 | 721 | 772 | 270 | 321 | bifunctional uroporphyrinogen-III synthetase/uroporphyrin-III C-methyltransferase; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.77e-72 | 8 | 690 | 11 | 603 | |
| 6.19e-67 | 376 | 477 | 1 | 102 | |
| 5.25e-66 | 291 | 477 | 1 | 195 | |
| 7.31e-66 | 291 | 477 | 1 | 195 | |
| 7.31e-66 | 291 | 477 | 1 | 195 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.38e-08 | 497 | 672 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
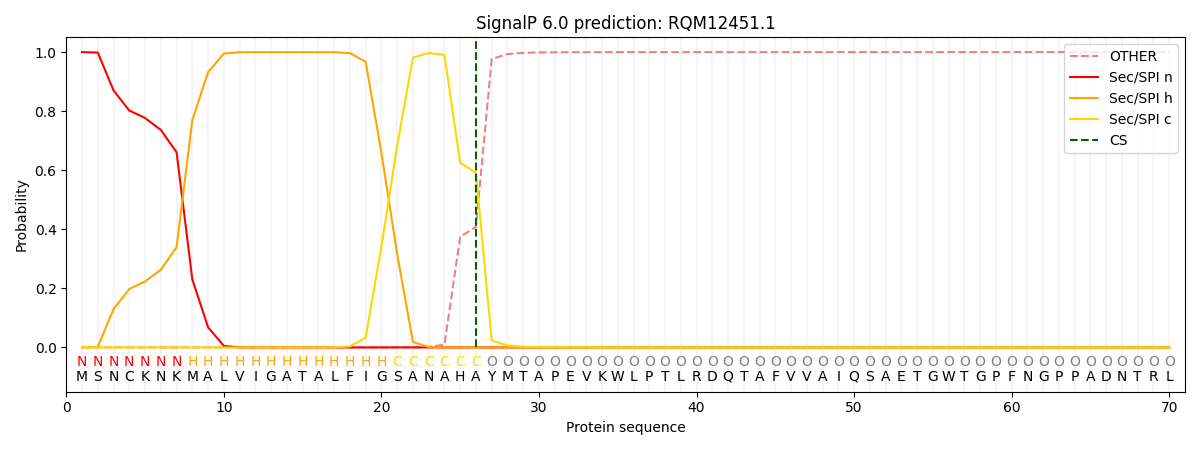
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000470 | 0.999485 | CS pos: 26-27. Pr: 0.5914 |

