You are browsing environment: FUNGIDB
CAZyme Information: RO3G_15845-t26_1-p1
Basic Information
help
| Species |
Rhizopus delemar
|
| Lineage |
Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar
|
| CAZyme ID |
RO3G_15845-t26_1-p1
|
| CAZy Family |
GT49 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 541 |
CH476748|CGC2 |
59879.03 |
4.6660 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_RdelemarRA99-880 |
17703 |
246409 |
244 |
17459
|
|
| Gene Location |
No EC number prediction in RO3G_15845-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA12 |
91 |
482 |
3.2e-36 |
0.9775561097256857 |
RO3G_15845-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 7.15e-27 |
105 |
480 |
68 |
458 |
| 1.80e-20 |
159 |
482 |
160 |
506 |
| 1.81e-20 |
159 |
482 |
161 |
507 |
| 2.55e-19 |
82 |
482 |
33 |
464 |
| 1.81e-18 |
100 |
482 |
39 |
419 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 3.27e-10 |
100 |
485 |
16 |
402 |
Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
| 3.28e-10 |
100 |
485 |
17 |
403 |
Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
| 3.29e-10 |
100 |
485 |
18 |
404 |
Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
RO3G_15845-t26_1-p1 has no Swissprot hit.
This protein is predicted as SP
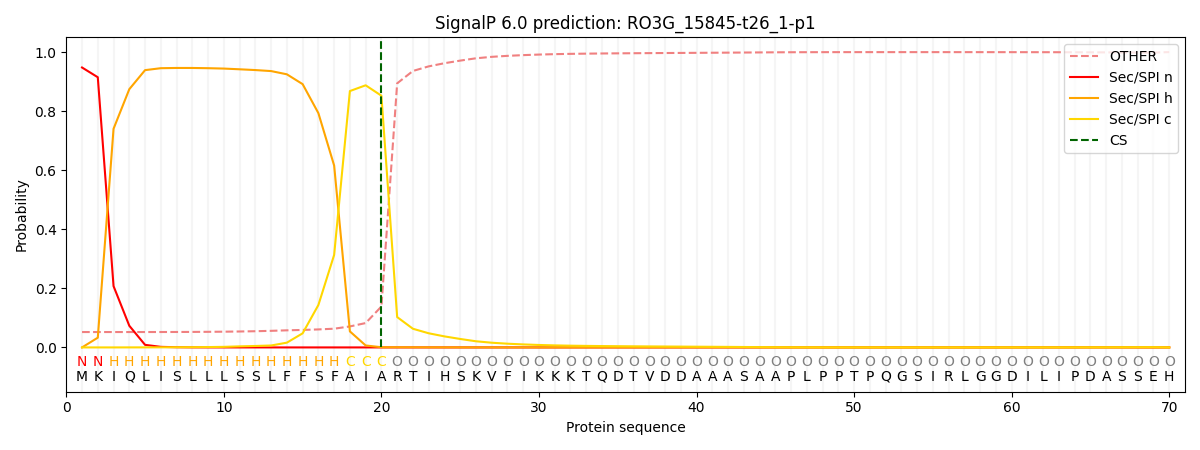
| Other |
SP_Sec_SPI |
CS Position |
| 0.061454 |
0.938523 |
CS pos: 20-21. Pr: 0.8523 |
There is no transmembrane helices in RO3G_15845-t26_1-p1.

