You are browsing environment: FUNGIDB
CAZyme Information: RO3G_15489-t26_1-p1
You are here: Home > Sequence: RO3G_15489-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_15489-t26_1-p1 | |||||||||||
| CAZy Family | GT48 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 42 | 676 | 8.9e-87 | 0.9664804469273743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 1.04e-76 | 22 | 690 | 3 | 530 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 177843 | PLN02191 | 4.10e-70 | 7 | 690 | 10 | 553 | L-ascorbate oxidase |
| 215324 | PLN02604 | 2.43e-68 | 5 | 690 | 9 | 553 | oxidoreductase |
| 274556 | laccase | 5.88e-52 | 18 | 681 | 1 | 518 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
| 259868 | CuRO_2_LCC_like | 5.43e-51 | 184 | 336 | 2 | 152 | Cupredoxin domain 2 of laccase-like multicopper oxidases; including laccase, CueO, spore coat protein A, ascorbate oxidase and similar proteins. Laccase-like multicopper oxidases (MCOs) are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs are capable of oxidizing a vast range of substrates, varying from aromatic compounds to inorganic compounds such as metals. Although the members of this family have diverse functions, majority of them have three cupredoxin domain repeats. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 2 of 3-domain MCOs has lost the ability to bind copper. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.25e-160 | 20 | 694 | 25 | 651 | |
| 5.62e-123 | 23 | 688 | 30 | 573 | |
| 9.55e-119 | 23 | 689 | 18 | 571 | |
| 2.61e-86 | 16 | 655 | 18 | 519 | |
| 1.73e-58 | 15 | 690 | 31 | 561 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.59e-45 | 20 | 690 | 3 | 530 | Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1AOZ_B Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1ASO_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASO_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASP_A X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASP_B X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASQ_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASQ_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo] |
|
| 2.56e-36 | 28 | 685 | 12 | 481 | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_B Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_C Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_D Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_E Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_F Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae] |
|
| 8.45e-29 | 23 | 676 | 6 | 523 | Crystal Structure of the Zea Mays laccase 3 [Zea mays],6KLI_A Crystal Structure of the Zea Mays laccase 3 complexed with sinapyl [Zea mays],6KLJ_A Crystal Structure of the Zea Mays laccase 3 complexed with coniferyl [Zea mays] |
|
| 3.34e-28 | 21 | 300 | 68 | 315 | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution [Botrytis aclada],4X4K_A Structure of laccase from Botrytis aclada with full copper content [Botrytis aclada] |
|
| 3.34e-28 | 21 | 300 | 68 | 315 | Structure of the L499M mutant of the laccase from B.aclada [Botrytis aclada] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.36e-52 | 7 | 690 | 7 | 549 | L-ascorbate oxidase OS=Brassica rapa subsp. pekinensis OX=51351 GN=AO PE=2 SV=1 |
|
| 3.27e-51 | 16 | 690 | 19 | 550 | L-ascorbate oxidase OS=Arabidopsis thaliana OX=3702 GN=AAO PE=1 SV=1 |
|
| 5.76e-48 | 7 | 688 | 22 | 564 | L-ascorbate oxidase OS=Cucumis sativus OX=3659 PE=1 SV=1 |
|
| 2.40e-47 | 18 | 690 | 28 | 557 | L-ascorbate oxidase OS=Nicotiana tabacum OX=4097 GN=AAO PE=2 SV=1 |
|
| 3.90e-44 | 20 | 690 | 3 | 530 | L-ascorbate oxidase OS=Cucurbita pepo var. melopepo OX=3665 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
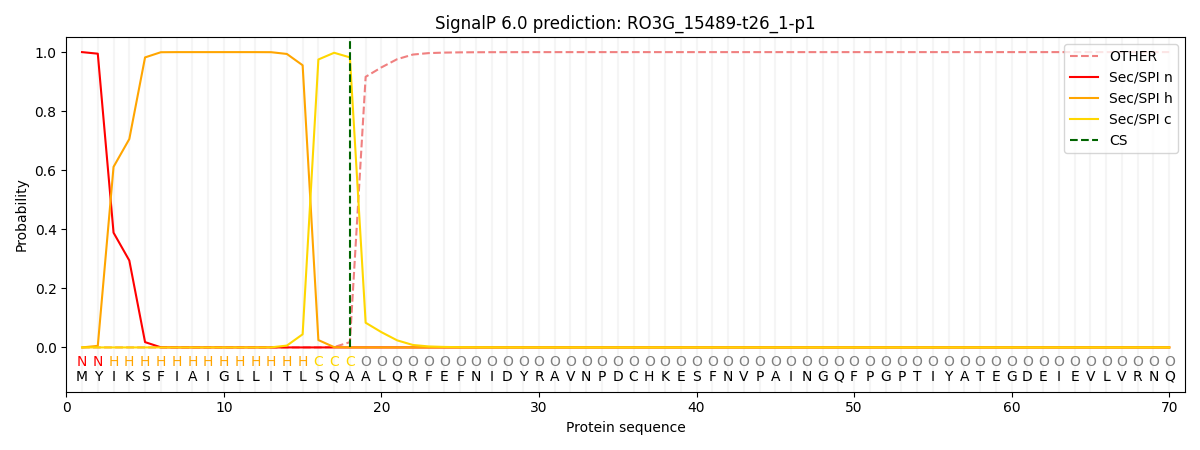
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000243 | 0.999717 | CS pos: 18-19. Pr: 0.9821 |
