You are browsing environment: FUNGIDB
RO3G_14582-t26_1-p1
Basic Information
help
Species
Rhizopus delemar
Lineage
Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar
CAZyme ID
RO3G_14582-t26_1-p1
CAZy Family
GT2|GT2
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
237
27571.91
9.3615
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_RdelemarRA99-880
17703
246409
244
17459
Gene Location
Family
Start
End
Evalue
family coverage
GT58
1
236
1.6e-96
0.6428571428571429
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
398745
ALG3
1.43e-133
1
237
29
264
ALG3 protein. The formation of N-glycosidic linkages of glycoproteins involves the ordered assembly of the common Glc3Man9GlcNAc2 core-oligosaccharide on the lipid carrier dolichyl pyrophosphate. Whereas early mannosylation steps occur on the cytoplasmic side of the endoplasmic reticulum with GDP-Man as donor, the final reactions from Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol on the lumenal side use Dol-P-Man. ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.68e-105
1
236
54
289
3.07e-72
1
234
69
303
3.07e-72
1
234
69
303
2.31e-71
1
237
71
308
8.71e-71
1
232
27
259
RO3G_14582-t26_1-p1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.15e-66
1
234
78
312
Lethal(2)neighbour of tid protein OS=Drosophila melanogaster OX=7227 GN=l(2)not PE=1 SV=1
more
1.76e-66
1
234
78
312
Lethal(2)neighbour of tid protein 2 OS=Drosophila melanogaster OX=7227 GN=l(2)not2 PE=2 SV=1
more
1.26e-65
1
234
77
311
Lethal(2)neighbour of Tid protein OS=Drosophila virilis OX=7244 GN=l(2)not PE=1 SV=1
more
1.90e-62
1
237
66
302
Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-3 PE=3 SV=1
more
4.14e-60
1
236
65
298
Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG3 PE=1 SV=1
more
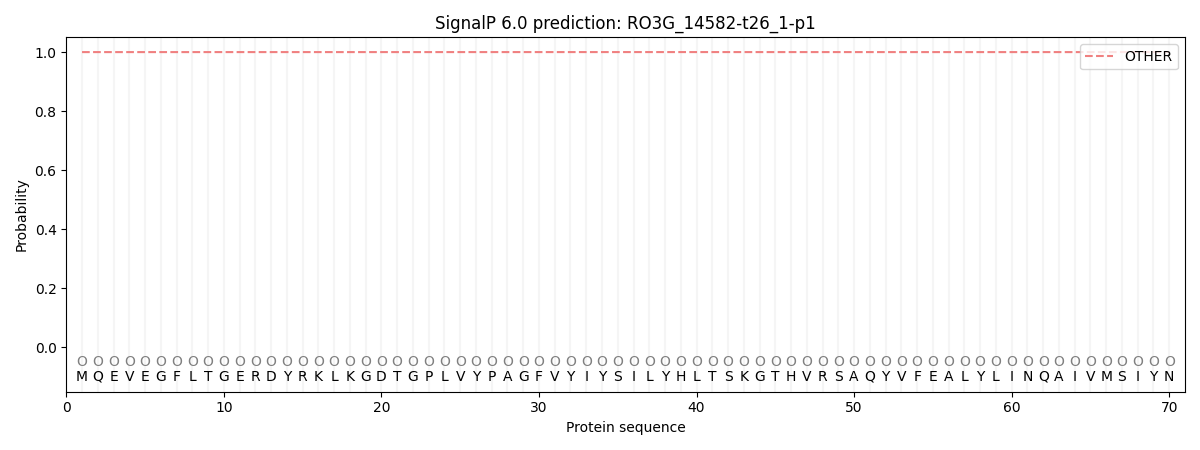
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000044
0.000001
Start
End
23
40
52
69
92
114
135
157
213
235