You are browsing environment: FUNGIDB
CAZyme Information: RO3G_10121-t26_1-p1
You are here: Home > Sequence: RO3G_10121-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_10121-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT77 | 130 | 334 | 1.9e-22 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403476 | Chs3p | 3.18e-149 | 343 | 613 | 5 | 283 | Chitin synthase III catalytic subunit. This family of proteins is found in eukaryotes. Proteins in this family are typically between 288 and 332 amino acids in length. This family is the catalytic domain of chitin synthase III. Chitin is a major component of fungal cell walls and this enzyme is responsible for its formation. |
| 367480 | Nucleotid_trans | 3.11e-13 | 126 | 336 | 1 | 208 | Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.57e-120 | 61 | 343 | 85 | 367 | |
| 8.75e-120 | 1 | 343 | 1 | 355 | |
| 9.55e-67 | 34 | 343 | 30 | 332 | |
| 6.61e-59 | 75 | 340 | 105 | 366 | |
| 3.17e-53 | 67 | 343 | 88 | 352 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.15e-105 | 338 | 639 | 8 | 316 | Chitin synthase export chaperone OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CHS7 PE=3 SV=1 |
|
| 3.23e-105 | 338 | 639 | 8 | 316 | Chitin synthase export chaperone OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=CHS7 PE=3 SV=1 |
|
| 3.07e-99 | 343 | 642 | 6 | 318 | Chitin synthase export chaperone OS=Coccidioides immitis (strain RS) OX=246410 GN=CHS7 PE=3 SV=1 |
|
| 8.34e-96 | 343 | 632 | 6 | 300 | Chitin synthase export chaperone OS=Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) OX=321614 GN=CHS7 PE=3 SV=1 |
|
| 1.00e-93 | 343 | 632 | 6 | 301 | Chitin synthase export chaperone OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHS7 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000010 | 0.000009 |
TMHMM Annotations download full data without filtering help
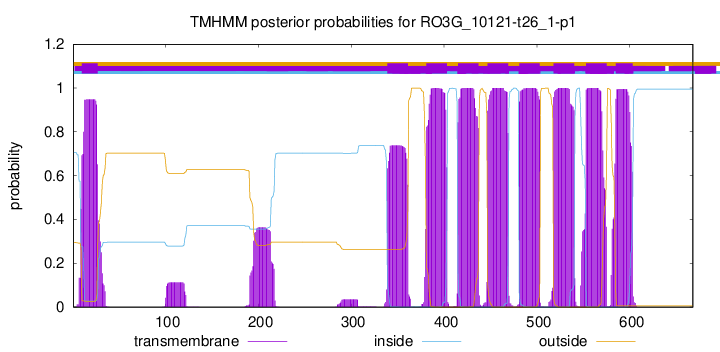
| Start | End |
|---|---|
| 10 | 27 |
| 339 | 361 |
| 381 | 403 |
| 415 | 437 |
| 447 | 469 |
| 481 | 503 |
| 518 | 540 |
| 553 | 575 |
| 585 | 603 |
