You are browsing environment: FUNGIDB
CAZyme Information: RO3G_07953-t26_1-p1
You are here: Home > Sequence: RO3G_07953-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
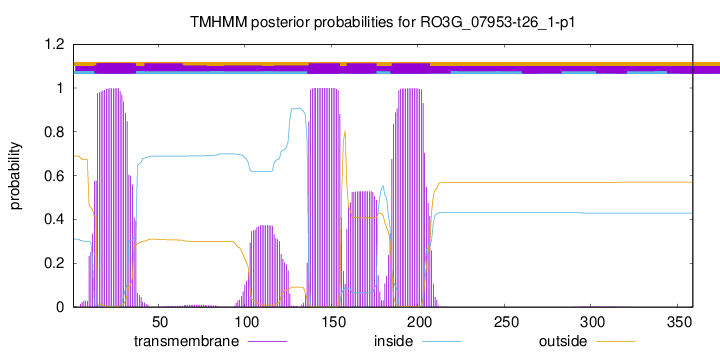
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_07953-t26_1-p1 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 5 | 224 | 6.6e-32 | 0.4910025706940874 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 9.91e-20 | 72 | 207 | 230 | 369 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.80e-84 | 9 | 354 | 105 | 483 | |
| 2.99e-24 | 37 | 210 | 209 | 376 | |
| 2.14e-23 | 56 | 223 | 219 | 386 | |
| 3.84e-23 | 46 | 211 | 218 | 375 | |
| 4.39e-23 | 46 | 211 | 219 | 376 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.83e-22 | 39 | 212 | 231 | 382 | GPI mannosyltransferase 4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=SMP3 PE=3 SV=2 |
|
| 9.03e-22 | 55 | 223 | 223 | 386 | GPI mannosyltransferase 4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SMP3 PE=1 SV=3 |
|
| 1.83e-21 | 56 | 210 | 228 | 374 | GPI mannosyltransferase 4 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=smp3 PE=3 SV=2 |
|
| 2.44e-21 | 49 | 210 | 226 | 379 | GPI mannosyltransferase 4 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=smp3 PE=3 SV=1 |
|
| 9.83e-21 | 58 | 223 | 226 | 382 | GPI mannosyltransferase 4 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=SMP3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
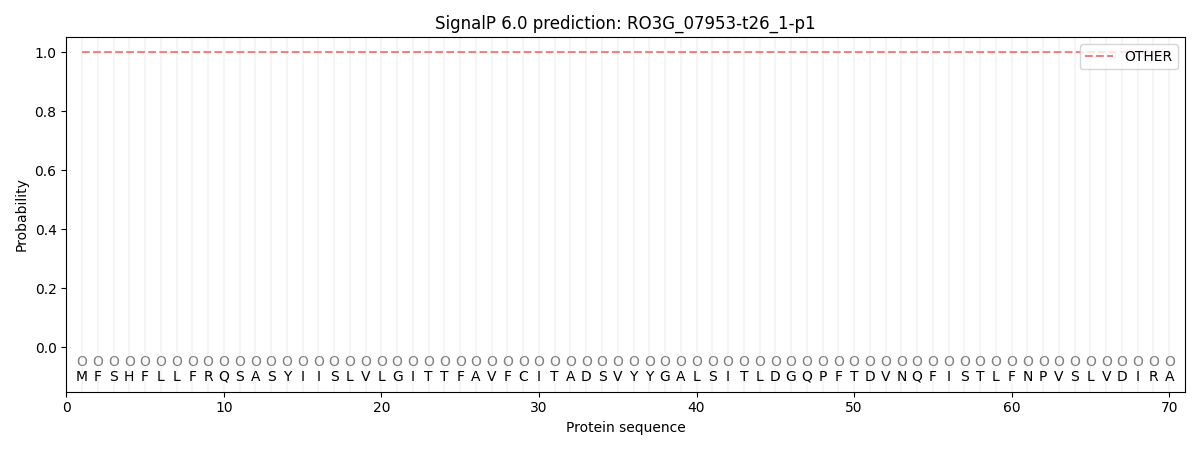
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000040 | 0.000015 |