You are browsing environment: FUNGIDB
CAZyme Information: RO3G_07453-t26_1-p1
You are here: Home > Sequence: RO3G_07453-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
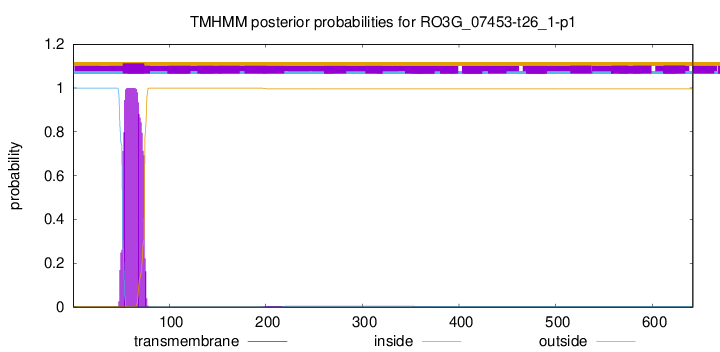
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_07453-t26_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT49 | 134 | 383 | 4.6e-28 | 0.7388724035608308 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403069 | Velvet | 5.72e-71 | 477 | 629 | 1 | 210 | Velvet factor. The velvet factor is conserved in many fungal species and is found to have gained different roles depending on the organism's need, expanding the conserved role in developmental programmes. The velvet factor orthologues can be adapted to the fungal-specific life cycle and may be involved in diverse functions such as sclerotia formation and toxin production, as in A. parasiticus, nutrition-dependent sporulation, as in A. fumigatus, or the microconidia-to-macroconidia ratio and cell wall formation, as in the heterothallic fungus Fusarium verticilloides. |
| 404735 | Glyco_transf_49 | 5.92e-25 | 140 | 395 | 2 | 317 | Glycosyl-transferase for dystroglycan. This glycosyl-transferase brings about the glycosylation of the alpha-dystroglycan subunit. Dystroglycan is an integral member of the skeletal muscular dystrophin glycoprotein complex, which links dystrophin to proteins in the extracellular matrix. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.38e-122 | 51 | 437 | 123 | 516 | |
| 5.93e-118 | 48 | 437 | 104 | 501 | |
| 6.19e-68 | 48 | 441 | 72 | 482 | |
| 2.25e-65 | 77 | 441 | 108 | 489 | |
| 1.42e-48 | 103 | 433 | 82 | 414 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.93e-21 | 477 | 624 | 20 | 156 | Crystal structure of VosA velvet domain [Aspergillus nidulans],4N6R_A Crystal structure of VosA-VelB-complex [Aspergillus nidulans] |
|
| 1.94e-19 | 475 | 629 | 57 | 321 | Crystal structure of VosA-VelB-complex [Aspergillus nidulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.98e-30 | 477 | 628 | 218 | 371 | Sexual development regulator VELC OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=VELC PE=3 SV=1 |
|
| 1.71e-25 | 477 | 628 | 244 | 397 | Sexual development regulator velC OS=Gibberella moniliformis (strain M3125 / FGSC 7600) OX=334819 GN=velC PE=1 SV=1 |
|
| 4.55e-23 | 476 | 628 | 74 | 228 | Spore development regulator vosA OS=Beauveria bassiana (strain ARSEF 2860) OX=655819 GN=vosA PE=2 SV=1 |
|
| 5.50e-21 | 462 | 629 | 189 | 364 | Sexual development regulator umv3 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=umv3 PE=3 SV=1 |
|
| 7.83e-20 | 473 | 629 | 56 | 316 | Velvet complex subunit B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=velB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
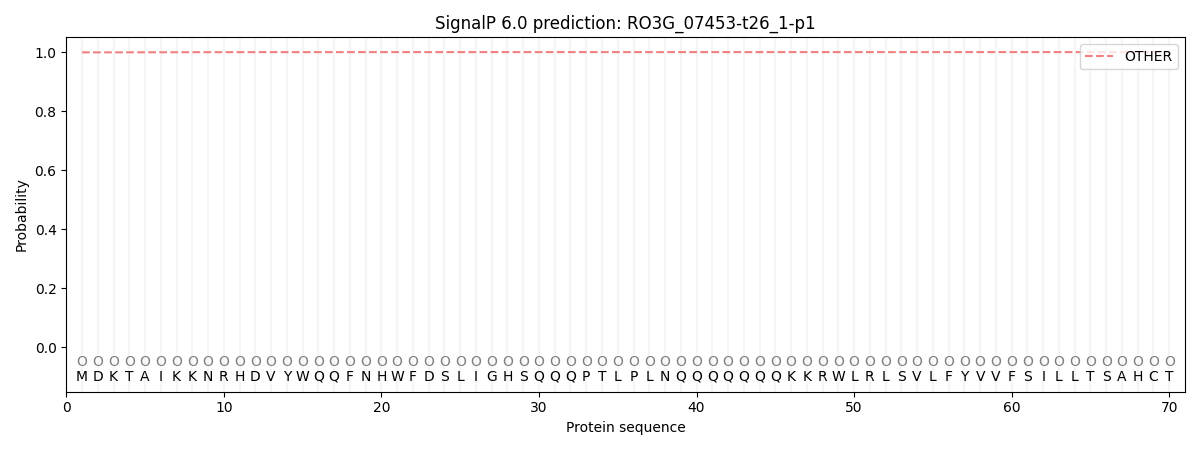
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999295 | 0.000689 |