You are browsing environment: FUNGIDB
CAZyme Information: RO3G_04907-t26_1-p1
You are here: Home > Sequence: RO3G_04907-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_04907-t26_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.149:9 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 9.95e-17 | 240 | 423 | 1 | 195 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 403517 | EOS1 | 1.36e-06 | 70 | 115 | 1 | 46 | N-glycosylation protein. This family is not required for survival of S.cerevisiae, but its deletion leads to heightened sensitivity to oxidative stress. It appears to be involved in N-glycosylation, and resides in the endoplasmic reticulum. |
| 398326 | Gb3_synth | 4.72e-06 | 716 | 818 | 5 | 106 | Alpha 1,4-glycosyltransferase conserved region. The glycosphingolipids (GSL) form part of eukaryotic cell membranes. They consist of a hydrophilic carbohydrate moiety linked to a hydrophobic ceramide tail embedded within the lipid bilayer of the membrane. Lactosylceramide, Gal1,4Glc1Cer (LacCer), is the common synthetic precursor to the majority of GSL found in vertebrates. Alpha 1.4-glycosyltransferases utilize UDP donors and transfer the sugar to a beta-linked acceptor. This region appears to be confined to higher eukaryotes. No function has been yet assigned to this region. |
| 398274 | Gly_transf_sug | 0.004 | 631 | 668 | 42 | 76 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 215596 | PLN03133 | 0.005 | 228 | 387 | 387 | 552 | beta-1,3-galactosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 3 | 899 | 2 | 907 | |
| 1.75e-174 | 228 | 897 | 292 | 978 | |
| 9.06e-93 | 7 | 506 | 7 | 516 | |
| 3.48e-18 | 538 | 829 | 210 | 483 | |
| 7.16e-18 | 549 | 826 | 617 | 887 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.31e-11 | 281 | 440 | 314 | 474 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 OS=Mus musculus OX=10090 GN=B3galnt2 PE=1 SV=1 |
|
| 5.53e-11 | 548 | 820 | 366 | 629 | Uncharacterized protein At4g19900 OS=Arabidopsis thaliana OX=3702 GN=At4g19900 PE=2 SV=1 |
|
| 1.56e-09 | 232 | 465 | 442 | 680 | Hydroxyproline O-galactosyltransferase GALT2 OS=Arabidopsis thaliana OX=3702 GN=GALT2 PE=1 SV=1 |
|
| 2.76e-08 | 229 | 421 | 153 | 356 | Beta-1,3-galactosyltransferase 2 OS=Homo sapiens OX=9606 GN=B3GALT2 PE=1 SV=1 |
|
| 2.76e-08 | 229 | 421 | 153 | 356 | Beta-1,3-galactosyltransferase 2 OS=Pongo abelii OX=9601 GN=B3GALT2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000032 | 0.000009 |
TMHMM Annotations download full data without filtering help
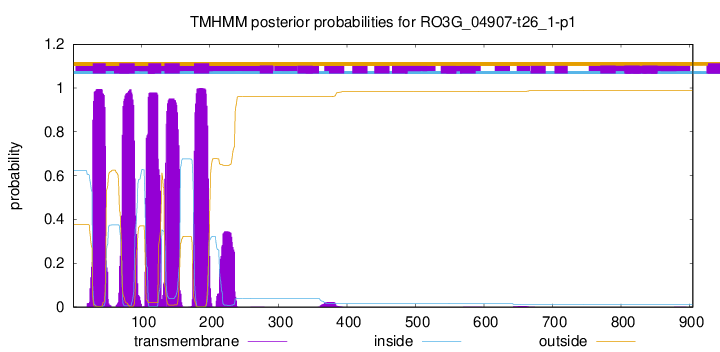
| Start | End |
|---|---|
| 29 | 48 |
| 68 | 90 |
| 110 | 129 |
| 134 | 156 |
| 177 | 199 |
