You are browsing environment: FUNGIDB
CAZyme Information: RO3G_04560-t26_1-p1
You are here: Home > Sequence: RO3G_04560-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
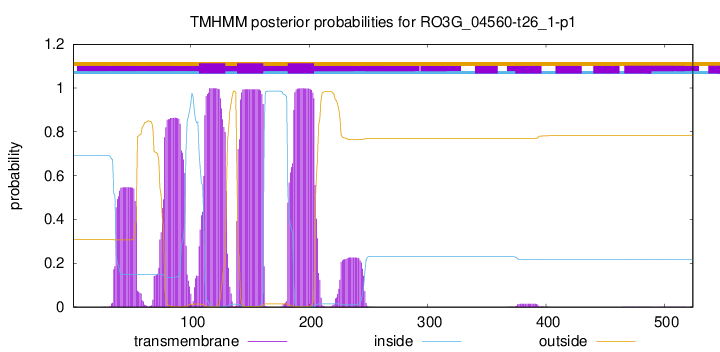
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_04560-t26_1-p1 | |||||||||||
| CAZy Family | GH15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403517 | EOS1 | 6.39e-16 | 76 | 166 | 1 | 93 | N-glycosylation protein. This family is not required for survival of S.cerevisiae, but its deletion leads to heightened sensitivity to oxidative stress. It appears to be involved in N-glycosylation, and resides in the endoplasmic reticulum. |
| 250845 | Galactosyl_T | 1.26e-13 | 248 | 434 | 1 | 196 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.80e-167 | 22 | 519 | 14 | 517 | |
| 2.19e-128 | 15 | 508 | 10 | 512 | |
| 9.70e-59 | 38 | 522 | 45 | 592 | |
| 1.20e-10 | 232 | 472 | 64 | 303 | |
| 1.10e-09 | 232 | 472 | 63 | 301 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.19e-07 | 198 | 472 | 339 | 617 | Hydroxyproline O-galactosyltransferase GALT3 OS=Arabidopsis thaliana OX=3702 GN=GALT3 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999946 | 0.000044 |