You are browsing environment: FUNGIDB
CAZyme Information: RO3G_04544-t26_1-p1
You are here: Home > Sequence: RO3G_04544-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
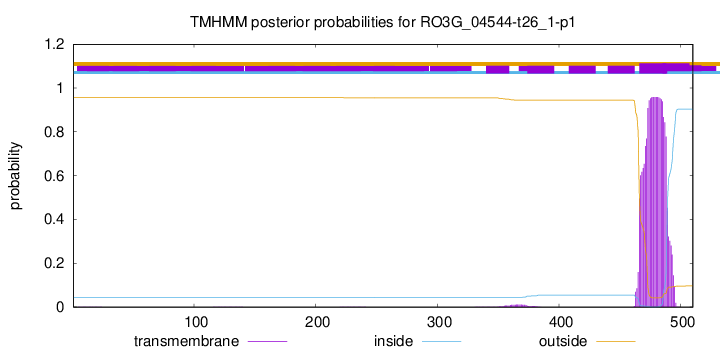
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_04544-t26_1-p1 | |||||||||||
| CAZy Family | GH134 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 220 | 443 | 1.2e-28 | 0.4476439790575916 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 1.19e-46 | 1 | 443 | 8 | 403 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 278624 | UDPGT | 5.60e-28 | 5 | 490 | 11 | 490 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 224732 | YjiC | 1.90e-23 | 2 | 452 | 10 | 405 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 273616 | MGT | 4.54e-21 | 11 | 442 | 13 | 387 | glycosyltransferase, MGT family. This model describes the MGT (macroside glycosyltransferase) subfamily of the UDP-glucuronosyltransferase family. Members include a number of glucosyl transferases for macrolide antibiotic inactivation, but also include transferases of glucose-related sugars for macrolide antibiotic production. [Cellular processes, Toxin production and resistance] |
| 223071 | egt | 7.86e-17 | 208 | 421 | 251 | 441 | ecdysteroid UDP-glucosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.92e-197 | 2 | 510 | 47 | 548 | |
| 1.73e-96 | 3 | 491 | 44 | 528 | |
| 8.85e-93 | 1 | 485 | 54 | 531 | |
| 2.03e-85 | 2 | 474 | 51 | 515 | |
| 3.89e-81 | 1 | 474 | 48 | 514 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.11e-13 | 213 | 383 | 210 | 385 | Structure of UGT78G1 complexed with myricetin and UDP [Medicago truncatula],3HBJ_A Structure of UGT78G1 complexed with UDP [Medicago truncatula] |
|
| 1.17e-12 | 250 | 428 | 264 | 442 | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea [Clitoria ternatea],4REL_A Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol [Clitoria ternatea],4REM_A Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin [Clitoria ternatea],4REN_A Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin [Clitoria ternatea],4WHM_A Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP [Clitoria ternatea] |
|
| 2.24e-12 | 326 | 445 | 280 | 394 | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_B Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_C Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_D Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora] |
|
| 2.27e-12 | 326 | 445 | 283 | 397 | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_B Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_C Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_D Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora] |
|
| 6.67e-11 | 326 | 425 | 69 | 167 | Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens],2O6L_B Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.85e-17 | 5 | 477 | 34 | 497 | UDP-glucuronosyltransferase 2B9 OS=Macaca fascicularis OX=9541 GN=UGT2B9 PE=2 SV=1 |
|
| 8.92e-17 | 5 | 477 | 34 | 497 | UDP-glucuronosyltransferase 2B33 OS=Macaca mulatta OX=9544 GN=UGT2B33 PE=1 SV=1 |
|
| 3.71e-16 | 5 | 479 | 34 | 499 | UDP-glucuronosyltransferase 2B7 OS=Homo sapiens OX=9606 GN=UGT2B7 PE=1 SV=2 |
|
| 8.71e-16 | 5 | 477 | 34 | 497 | UDP-glucuronosyltransferase 2B18 OS=Macaca fascicularis OX=9541 GN=UGT2B18 PE=1 SV=1 |
|
| 8.74e-16 | 5 | 477 | 34 | 498 | UDP-glucuronosyltransferase 2A3 OS=Cavia porcellus OX=10141 GN=UGT2A3 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
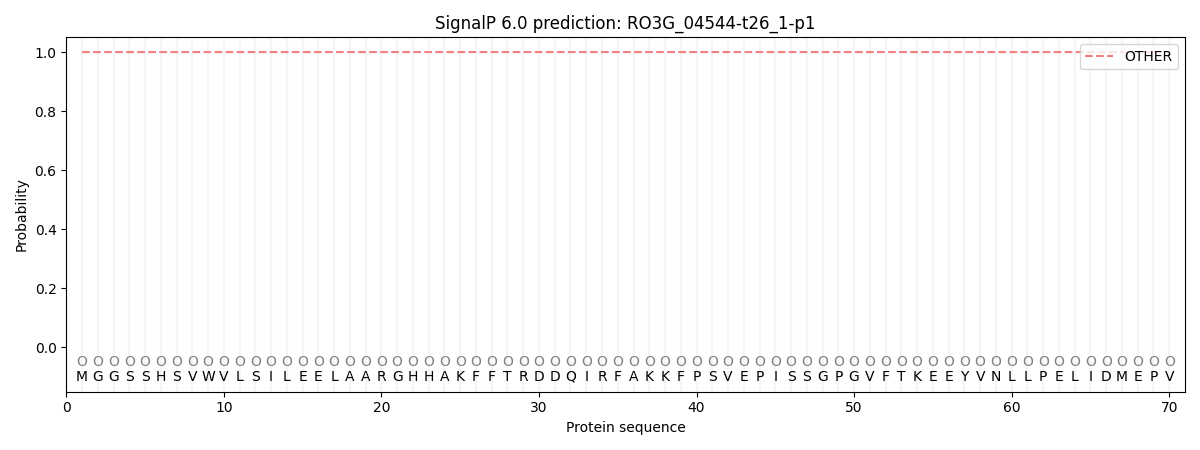
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000078 | 0.000000 |