You are browsing environment: FUNGIDB
CAZyme Information: RO3G_01554-t26_1-p1
You are here: Home > Sequence: RO3G_01554-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus delemar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus delemar | |||||||||||
| CAZyme ID | RO3G_01554-t26_1-p1 | |||||||||||
| CAZy Family | CE20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.26:2 |
|---|
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.46e-89 | 33 | 303 | 27 | 289 | |
| 2.87e-85 | 32 | 298 | 24 | 283 | |
| 6.18e-57 | 76 | 303 | 60 | 283 | |
| 1.21e-54 | 76 | 303 | 174 | 392 | |
| 1.23e-51 | 78 | 298 | 46 | 259 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.22e-43 | 75 | 299 | 39 | 269 | Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai] |
|
| 9.94e-17 | 94 | 301 | 39 | 241 | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3GNE_A Crystal structure of alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3GNE_B Crystal structure of alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3IM0_A Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) [Paramecium bursaria Chlorella virus CVK2] |
|
| 1.12e-10 | 113 | 297 | 56 | 234 | Structure of alginate lyase Aly36B [Chitinophaga sp. MD30] |
|
| 4.14e-09 | 113 | 297 | 56 | 234 | Structure of alginate lyase Aly36B mutant K143A/M171A in complex with alginate trisaccharide [Chitinophaga sp. MD30],6KZK_B Structure of alginate lyase Aly36B mutant K143A/M171A in complex with alginate trisaccharide [Chitinophaga sp. MD30],6KZK_C Structure of alginate lyase Aly36B mutant K143A/M171A in complex with alginate trisaccharide [Chitinophaga sp. MD30] |
|
| 1.05e-08 | 113 | 297 | 56 | 234 | Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide [Chitinophaga sp. MD30],6KCV_B Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide [Chitinophaga sp. MD30],6KCV_C Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide [Chitinophaga sp. MD30] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
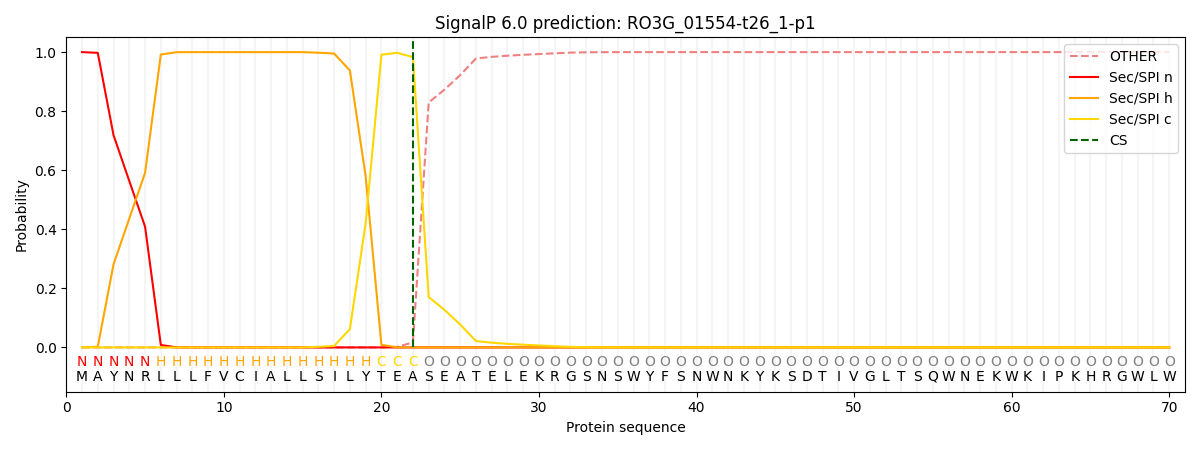
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000201 | 0.999772 | CS pos: 22-23. Pr: 0.9830 |
