You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_09999-RA-p1
You are here: Home > Sequence: RL4_JR_09999-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
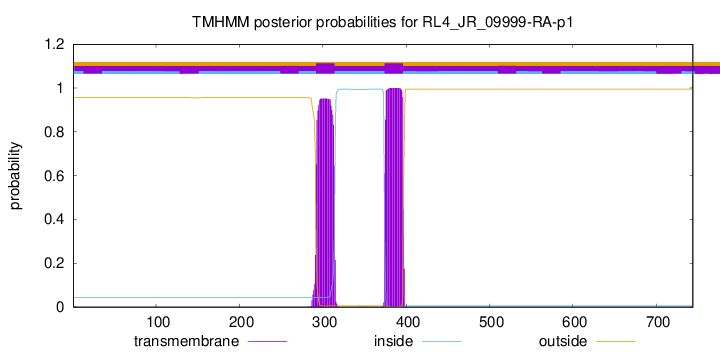
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_09999-RA-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 463 | 730 | 8.5e-32 | 0.9453376205787781 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.13e-33 | 444 | 727 | 45 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 3.48e-12 | 467 | 730 | 25 | 304 | Glycosyl hydrolases family 17. |
| 409231 | KLF1_2_4_N-like | 0.007 | 20 | 77 | 219 | 278 | N-terminal domain of Kruppel-like factors with similarity to the N-terminal domains of Kruppel-like factor (KLF)1, KLF2, and KLF4. Kruppel/Krueppel-like transcription factors (KLFs) belong to a family of proteins called the Specificity Protein (SP)/KLF family, characterized by a C-terminal DNA-binding domain of 81 amino acids consisting of three Kruppel-like C2H2 zinc fingers. These factors bind to a loose consensus motif, namely NNRCRCCYY (where N is any nucleotide; R is A/G, and Y is C/T), such as the recurring motifs in GC and GT boxes (5'-GGGGCGGGG-3' and 5-GGTGTGGGG-3') that are present in promoters and more distal regulatory elements of mammalian genes. Members of the KLF family can act as activators or repressors of transcription depending on cell and promoter context. KLFs regulate various cellular functions, such as proliferation, differentiation, and apoptosis, as well as the development and homeostasis of several types of tissue. In addition to the C-terminal DNA-binding domain, each KLF also has a unique N-terminal activation/repression domain that confers specifity and allows it to bind specifically to a certain partner, leading to distinct activities in vivo. This model represents the N-terminal domains of an unknown subfamily of KLFs, predominantly found in fish, related to the N-terminal domains of KLF1, KLF2, and KLF4. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.38e-211 | 2 | 741 | 4 | 766 | |
| 1.10e-208 | 1 | 742 | 1 | 729 | |
| 8.40e-208 | 61 | 744 | 36 | 718 | |
| 8.40e-208 | 61 | 744 | 36 | 718 | |
| 2.38e-207 | 61 | 744 | 36 | 718 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.86e-24 | 461 | 735 | 47 | 294 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 6.03e-23 | 461 | 735 | 47 | 294 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.60e-146 | 130 | 744 | 82 | 685 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgC PE=3 SV=1 |
|
| 5.60e-146 | 130 | 744 | 82 | 685 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgC PE=3 SV=2 |
|
| 6.14e-143 | 133 | 743 | 56 | 654 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgC PE=3 SV=2 |
|
| 3.70e-141 | 133 | 743 | 80 | 687 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgC PE=3 SV=1 |
|
| 1.01e-140 | 119 | 743 | 66 | 687 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
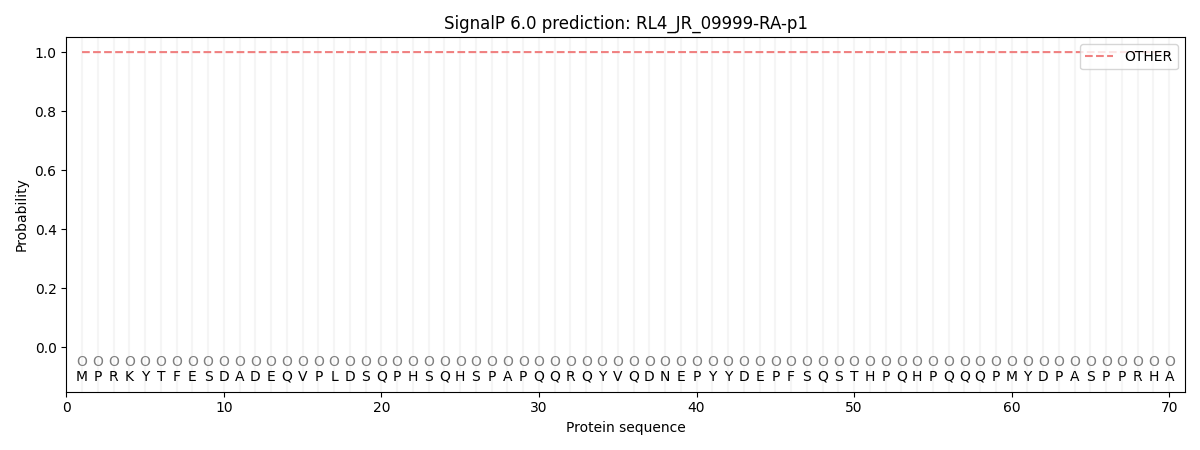
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000027 | 0.000000 |