You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_09409-RA-p1
You are here: Home > Sequence: RL4_JR_09409-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_09409-RA-p1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 47 | 273 | 1.6e-67 | 0.9776785714285714 |
| GT2 | 959 | 1192 | 2.4e-50 | 0.9898477157360406 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 371727 | Glyco_hydro_cc | 5.54e-79 | 37 | 273 | 3 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| 206738 | DLP_1 | 1.43e-50 | 354 | 641 | 5 | 273 | Dynamin_like protein family includes dynamins and Mx proteins. The dynamin family of large mechanochemical GTPases includes the classical dynamins and dynamin-like proteins (DLPs) that are found throughout the Eukarya. These proteins catalyze membrane fission during clathrin-mediated endocytosis. Dynamin consists of five domains; an N-terminal G domain that binds and hydrolyzes GTP, a middle domain (MD) involved in self-assembly and oligomerization, a pleckstrin homology (PH) domain responsible for interactions with the plasma membrane, GED, which is also involved in self-assembly, and a proline arginine rich domain (PRD) that interacts with SH3 domains on accessory proteins. To date, three vertebrate dynamin genes have been identified; dynamin 1, which is brain specific, mediates uptake of synaptic vesicles in presynaptic terminals; dynamin-2 is expressed ubiquitously and similarly participates in membrane fission; mutations in the MD, PH and GED domains of dynamin 2 have been linked to human diseases such as Charcot-Marie-Tooth peripheral neuropathy and rare forms of centronuclear myopathy. Dynamin 3 participates in megakaryocyte progenitor amplification, and is also involved in cytoplasmic enlargement and the formation of the demarcation membrane system. This family also includes interferon-induced Mx proteins that inhibit a wide range of viruses by blocking an early stage of the replication cycle. Dynamin oligomerizes into helical structures around the neck of budding vesicles in a GTP hydrolysis-dependent manner. |
| 404513 | Glyco_trans_2_3 | 2.81e-46 | 959 | 1193 | 1 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 197491 | DYNc | 1.02e-17 | 355 | 595 | 29 | 239 | Dynamin, GTPase. Large GTPases that mediate vesicle trafficking. Dynamin participates in the endocytic uptake of receptors, associated ligands, and plasma membrane following an exocytic event. |
| 395279 | Dynamin_N | 5.97e-12 | 355 | 552 | 1 | 167 | Dynamin family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 347 | 1360 | 19 | 1016 | |
| 0.0 | 347 | 1360 | 19 | 1034 | |
| 0.0 | 347 | 1360 | 19 | 1034 | |
| 0.0 | 460 | 1360 | 6 | 912 | |
| 2.51e-312 | 762 | 1360 | 52 | 642 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.40e-23 | 355 | 684 | 37 | 345 | GDP-bound stalkless-MxA [Homo sapiens] |
|
| 3.16e-21 | 355 | 684 | 37 | 348 | Nucleotide-free stalkless-MxA [Homo sapiens] |
|
| 3.30e-21 | 355 | 649 | 31 | 305 | GMPPCP-bound stalkless-MxA [Homo sapiens] |
|
| 5.32e-21 | 355 | 644 | 16 | 285 | GMPPCP-bound stalkless-MxA [Homo sapiens] |
|
| 1.20e-19 | 355 | 649 | 41 | 315 | Modified human MxA, nucleotide-free form [Homo sapiens],5GTM_B Modified human MxA, nucleotide-free form [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.90e-24 | 25 | 274 | 294 | 529 | Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1 |
|
| 1.86e-20 | 355 | 667 | 37 | 341 | Interferon-induced GTP-binding protein Mx1 OS=Ictalurus punctatus OX=7998 GN=mx1 PE=2 SV=1 |
|
| 5.50e-20 | 355 | 647 | 46 | 325 | Interferon-induced GTP-binding protein MxC OS=Danio rerio OX=7955 GN=mxc PE=2 SV=1 |
|
| 1.35e-19 | 355 | 666 | 40 | 334 | Interferon-induced GTP-binding protein MxA OS=Danio rerio OX=7955 GN=mxa PE=2 SV=1 |
|
| 1.58e-19 | 355 | 612 | 121 | 355 | Interferon-induced GTP-binding protein Mx2 OS=Canis lupus familiaris OX=9615 GN=MX2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
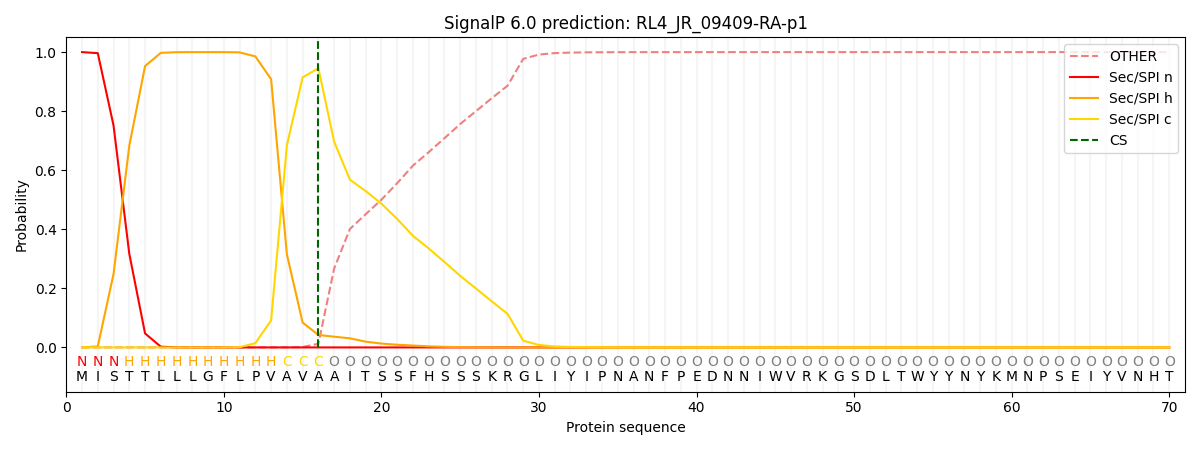
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000697 | 0.999281 | CS pos: 16-17. Pr: 0.9452 |
TMHMM Annotations download full data without filtering help
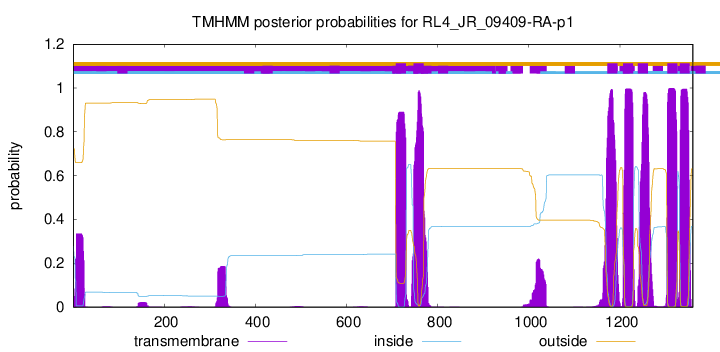
| Start | End |
|---|---|
| 708 | 730 |
| 747 | 769 |
| 1172 | 1194 |
| 1207 | 1229 |
| 1239 | 1261 |
| 1304 | 1326 |
| 1331 | 1353 |
