You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_08504-RA-p1
You are here: Home > Sequence: RL4_JR_08504-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_08504-RA-p1 | |||||||||||
| CAZy Family | GH92 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 17 | 197 | 9e-31 | 0.5407166123778502 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 407954 | DUF5597 | 3.57e-34 | 387 | 517 | 2 | 130 | Domain of unknown function (DUF5597). This is the C-terminal domain of xyloglucan utilization locus (XyGUL) present in Cellvibrio japonicas. XyGUL is required for xyloglucan utilization. It is also the C-terminal domain of PF02449 and PF01301. |
| 396048 | Glyco_hydro_35 | 9.59e-08 | 16 | 199 | 1 | 171 | Glycosyl hydrolases family 35. |
| 224786 | GanA | 1.11e-07 | 16 | 366 | 7 | 407 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 226217 | XynA | 7.97e-05 | 20 | 216 | 31 | 189 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 2.16e-04 | 49 | 195 | 19 | 154 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.38e-298 | 3 | 554 | 14 | 563 | |
| 1.54e-297 | 4 | 554 | 11 | 566 | |
| 6.22e-288 | 3 | 554 | 8 | 558 | |
| 6.22e-288 | 5 | 554 | 10 | 558 | |
| 3.58e-287 | 5 | 554 | 10 | 558 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.19e-78 | 11 | 513 | 32 | 516 | Chain A, Beta-galactosidase, GH35 family [Xanthomonas citri pv. citri str. 306],7KMO_A Chain A, Beta-galactosidase, GH35 [Xanthomonas citri pv. citri str. 306] |
|
| 7.82e-76 | 2 | 513 | 2 | 520 | The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_B The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_C The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_D The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_E The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_F The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_G The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1I_H The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],4D1J_A The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_B The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_C The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_D The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_E The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_F The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_G The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107],4D1J_H The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin [Cellvibrio japonicus Ueda107] |
|
| 9.94e-76 | 2 | 513 | 12 | 530 | Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_B Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_C Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_D Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_E Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_F Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_G Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],5JAW_H Structure of a beta galactosidase with inhibitor [Cellvibrio japonicus Ueda107],6TBF_A Chain A, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_B Chain B, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_C Chain C, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_D Chain D, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_E Chain E, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_F Chain F, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_G Chain G, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBF_H Chain H, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_A Chain A, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_B Chain B, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_C Chain C, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_D Chain D, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_E Chain E, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_F Chain F, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_G Chain G, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBG_H Chain H, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_A Chain A, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_B Chain B, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_C Chain C, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_D Chain D, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_E Chain E, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_F Chain F, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_G Chain G, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBH_H Chain H, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBI_A Chain A, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_B Chain B, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_C Chain C, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_D Chain D, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_E Chain E, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_F Chain F, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_G Chain G, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBI_H Chain H, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus],6TBJ_A Chain A, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_B Chain B, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_C Chain C, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_D Chain D, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_E Chain E, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_F Chain F, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_G Chain G, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBJ_H Chain H, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_A Chain A, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_B Chain B, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_C Chain C, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_D Chain D, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_E Chain E, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_F Chain F, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_G Chain G, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107],6TBK_H Chain H, Beta-galactosidase, putative, bgl35A [Cellvibrio japonicus Ueda107] |
|
| 1.26e-71 | 11 | 513 | 45 | 535 | The structure of a putative Beta-galactosidase from Caulobacter crescentus CB15. [Caulobacter vibrioides NA1000] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
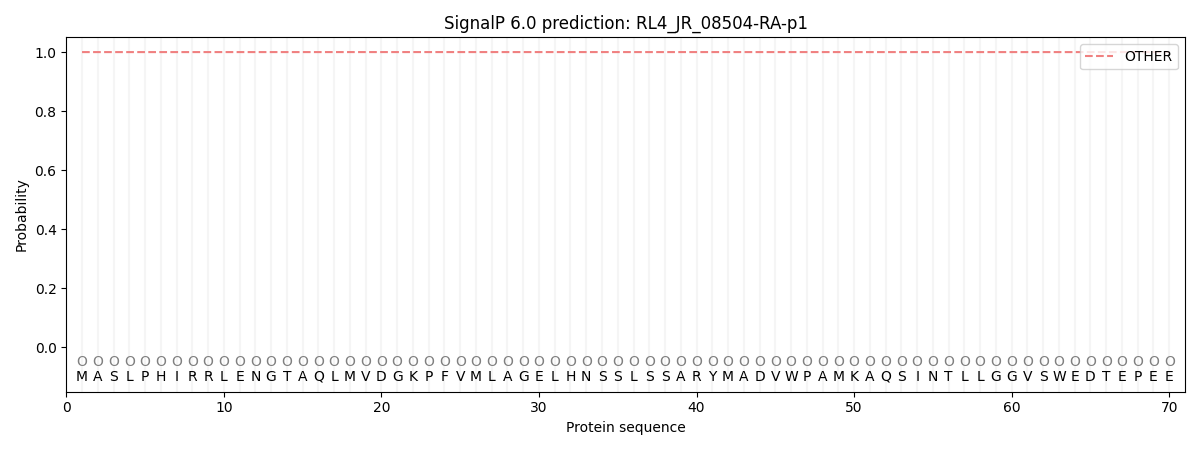
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000001 |
