You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_06216-RA-p1
You are here: Home > Sequence: RL4_JR_06216-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
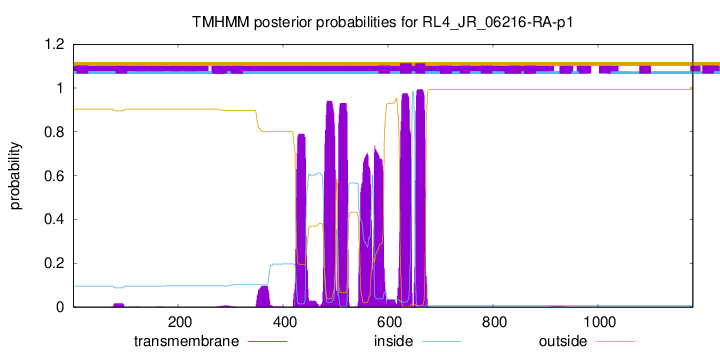
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_06216-RA-p1 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404385 | Methyltransf_23 | 3.74e-16 | 104 | 252 | 25 | 161 | Methyltransferase domain. This family appears to be a methyltransferase domain. |
| 404528 | Methyltransf_25 | 6.68e-13 | 105 | 192 | 1 | 97 | Methyltransferase domain. This family appears to be a methyltransferase domain. |
| 214773 | CAP10 | 5.14e-10 | 1063 | 1166 | 150 | 253 | Putative lipopolysaccharide-modifying enzyme. |
| 100107 | AdoMet_MTases | 9.34e-08 | 104 | 193 | 1 | 99 | S-adenosylmethionine-dependent methyltransferases (SAM or AdoMet-MTase), class I; AdoMet-MTases are enzymes that use S-adenosyl-L-methionine (SAM or AdoMet) as a substrate for methyltransfer, creating the product S-adenosyl-L-homocysteine (AdoHcy). There are at least five structurally distinct families of AdoMet-MTases, class I being the largest and most diverse. Within this class enzymes can be classified by different substrate specificities (small molecules, lipids, nucleic acids, etc.) and different target atoms for methylation (nitrogen, oxygen, carbon, sulfur, etc.). |
| 310354 | Glyco_transf_90 | 8.29e-07 | 1060 | 1173 | 217 | 332 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.26e-257 | 360 | 1172 | 11 | 829 | |
| 6.17e-250 | 362 | 1176 | 13 | 831 | |
| 6.14e-243 | 360 | 1172 | 9 | 821 | |
| 2.00e-241 | 360 | 1170 | 11 | 820 | |
| 1.16e-240 | 361 | 1173 | 12 | 823 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.21e-70 | 37 | 335 | 14 | 324 | Methyltransferase pytC OS=Aspergillus terreus OX=33178 GN=pytC PE=2 SV=1 |
|
| 1.08e-69 | 37 | 335 | 14 | 324 | Methyltransferase pytC OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pytC PE=2 SV=2 |
|
| 1.23e-46 | 58 | 334 | 14 | 305 | Secondary metabolism regulator LAE1 OS=Cochliobolus heterostrophus (strain C5 / ATCC 48332 / race O) OX=701091 GN=LAE1 PE=3 SV=1 |
|
| 3.81e-44 | 56 | 334 | 60 | 350 | Methyltransferase FFUJ_09178 OS=Gibberella fujikuroi (strain CBS 195.34 / IMI 58289 / NRRL A-6831) OX=1279085 GN=FFUJ_09178 PE=2 SV=1 |
|
| 4.46e-39 | 71 | 322 | 43 | 293 | Probable methyltransferase tdiE OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=tdiE PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000058 | 0.000001 |