You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_02015-RA-p1
You are here: Home > Sequence: RL4_JR_02015-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_02015-RA-p1 | |||||||||||
| CAZy Family | AA8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 26 | 340 | 1.2e-69 | 0.8798882681564246 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 5.48e-35 | 36 | 315 | 13 | 315 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170 | COG4833 | 8.59e-13 | 52 | 320 | 46 | 313 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| 395616 | Glyco_hydro_9 | 4.09e-08 | 71 | 192 | 187 | 292 | Glycosyl hydrolase family 9. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.47e-164 | 1 | 362 | 1 | 358 | |
| 4.73e-148 | 1 | 362 | 1 | 357 | |
| 5.78e-143 | 36 | 362 | 1 | 323 | |
| 1.73e-120 | 7 | 362 | 13 | 376 | |
| 1.97e-116 | 7 | 362 | 12 | 374 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.92e-11 | 84 | 314 | 103 | 324 | Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
|
| 3.58e-10 | 53 | 326 | 39 | 316 | Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 3.63e-10 | 53 | 326 | 42 | 319 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
|
| 3.92e-10 | 53 | 326 | 60 | 337 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 6.03e-10 | 53 | 326 | 73 | 350 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.95e-12 | 36 | 239 | 42 | 260 | Putative mannan endo-1,6-alpha-mannosidase C1198.06c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.06c PE=3 SV=1 |
|
| 1.10e-08 | 102 | 238 | 122 | 272 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
|
| 2.54e-08 | 52 | 240 | 65 | 269 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DCW1 PE=1 SV=1 |
|
| 1.04e-06 | 39 | 238 | 63 | 269 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=DCW1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
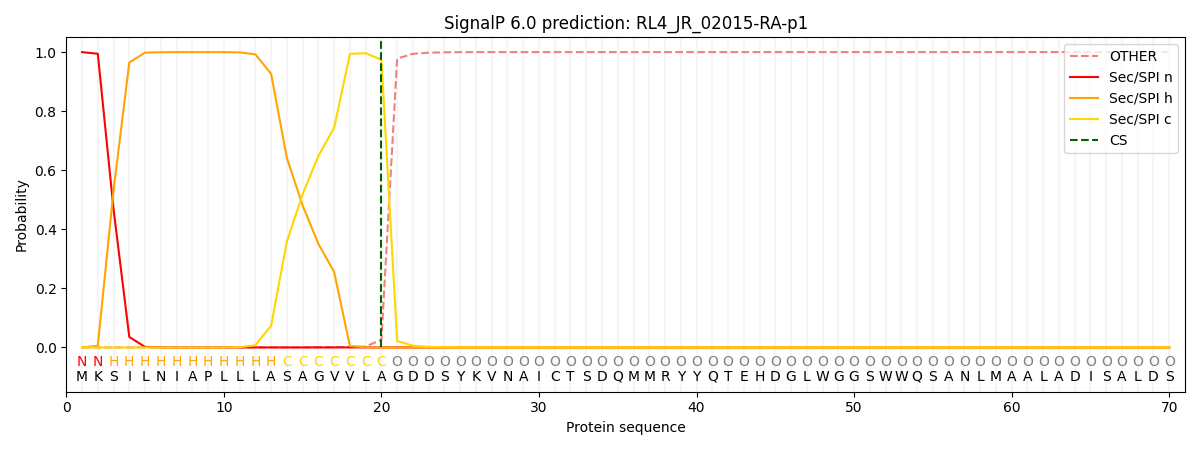
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000274 | 0.999694 | CS pos: 20-21. Pr: 0.9741 |
