You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_00890-RA-p1
You are here: Home > Sequence: RL4_JR_00890-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_00890-RA-p1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.55:10 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214830 | Alpha-L-AF_C | 2.70e-31 | 457 | 625 | 17 | 188 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| 399741 | Alpha-L-AF_C | 5.80e-28 | 459 | 625 | 23 | 191 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| 226064 | AbfA | 4.39e-23 | 201 | 633 | 33 | 499 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.68e-266 | 18 | 632 | 22 | 627 | |
| 7.93e-264 | 19 | 634 | 22 | 637 | |
| 3.70e-262 | 19 | 634 | 22 | 637 | |
| 5.91e-256 | 18 | 632 | 18 | 625 | |
| 3.07e-251 | 20 | 632 | 20 | 616 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.11e-127 | 21 | 632 | 1 | 624 | Chain AAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.39e-212 | 15 | 630 | 20 | 625 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=abfA PE=3 SV=1 |
|
| 1.78e-205 | 15 | 630 | 20 | 625 | Alpha-L-arabinofuranosidase A OS=Aspergillus niger OX=5061 GN=abfA PE=1 SV=1 |
|
| 3.58e-205 | 15 | 630 | 20 | 625 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=abfA PE=3 SV=1 |
|
| 2.11e-204 | 15 | 630 | 20 | 626 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
|
| 8.21e-204 | 15 | 630 | 20 | 625 | Alpha-L-arabinofuranosidase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=abfA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
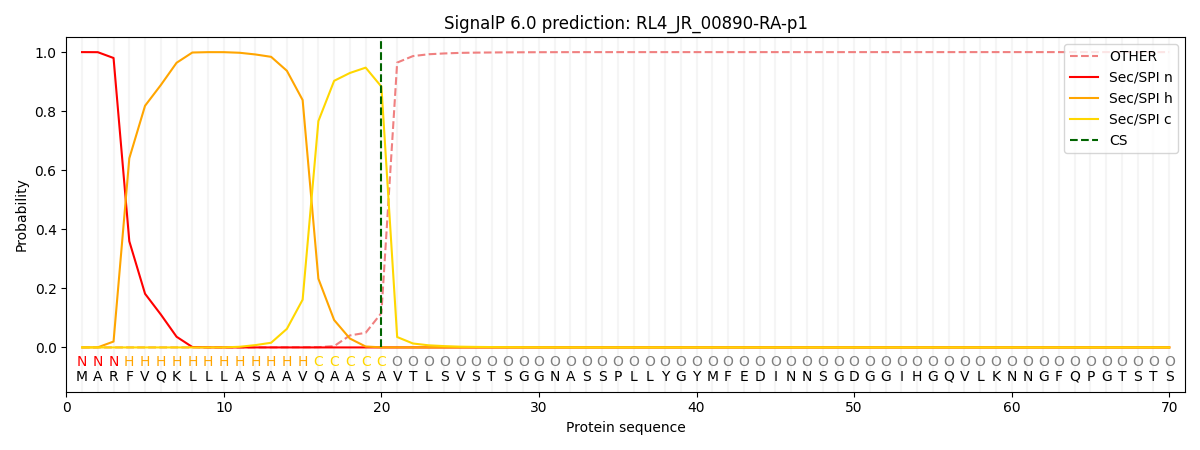
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000325 | 0.999640 | CS pos: 20-21. Pr: 0.8830 |
