You are browsing environment: FUNGIDB
CAZyme Information: RL4_JR_00288-RA-p1
You are here: Home > Sequence: RL4_JR_00288-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Raffaelea lauricola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Raffaelea; Raffaelea lauricola | |||||||||||
| CAZyme ID | RL4_JR_00288-RA-p1 | |||||||||||
| CAZy Family | AA12 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 8 | 410 | 1.5e-84 | 0.9203084832904884 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 4.25e-20 | 138 | 410 | 113 | 378 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.01e-179 | 1 | 600 | 2 | 592 | |
| 9.36e-176 | 1 | 595 | 1 | 567 | |
| 3.94e-174 | 12 | 598 | 5 | 584 | |
| 2.53e-171 | 2 | 601 | 37 | 650 | |
| 2.63e-164 | 5 | 601 | 7 | 573 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.07e-51 | 11 | 437 | 16 | 405 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG12 PE=1 SV=1 |
|
| 1.22e-50 | 15 | 437 | 11 | 403 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Caenorhabditis elegans OX=6239 GN=algn-12 PE=1 SV=2 |
|
| 1.82e-49 | 15 | 504 | 13 | 466 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG12 PE=1 SV=1 |
|
| 3.64e-48 | 11 | 437 | 17 | 404 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Mus musculus OX=10090 GN=Alg12 PE=2 SV=2 |
|
| 4.15e-44 | 24 | 504 | 28 | 434 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg12 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
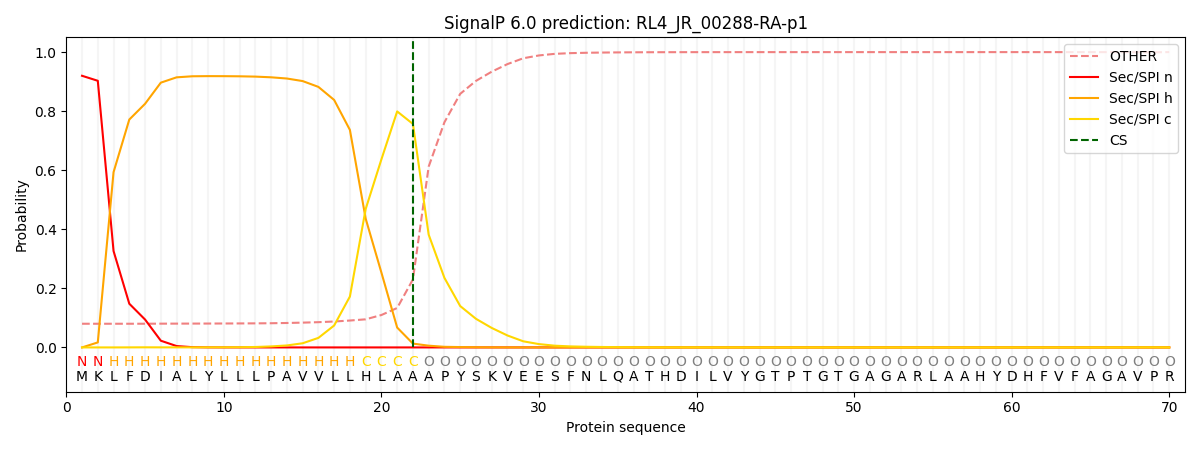
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.084555 | 0.915404 | CS pos: 22-23. Pr: 0.7567 |
TMHMM Annotations download full data without filtering help

| Start | End |
|---|---|
| 195 | 217 |
| 227 | 249 |
| 314 | 331 |
| 336 | 358 |
| 371 | 393 |
| 7 | 26 |
| 67 | 89 |
| 110 | 132 |
| 152 | 174 |
