You are browsing environment: FUNGIDB
CAZyme Information: RHZ58382.1
You are here: Home > Sequence: RHZ58382.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus thermomutatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus thermomutatus | |||||||||||
| CAZyme ID | RHZ58382.1 | |||||||||||
| CAZy Family | GH43|CBM91 | |||||||||||
| CAZyme Description | carbohydrate esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.72:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 27 | 232 | 1.7e-42 | 0.9947089947089947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 2.38e-61 | 27 | 232 | 1 | 173 | Cutinase. |
| 395595 | CBM_1 | 1.94e-11 | 254 | 282 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 2.38e-11 | 253 | 286 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 369775 | PE-PPE | 1.08e-04 | 60 | 133 | 1 | 76 | PE-PPE domain. This domain is found C terminal to the PE (pfam00934) and PPE (pfam00823) domains. The secondary structure of this domain is predicted to be a mixture of alpha helices and beta strands. |
| 235154 | PRK03739 | 0.009 | 61 | 99 | 508 | 548 | 2-isopropylmalate synthase; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.72e-138 | 5 | 286 | 9 | 300 | |
| 1.76e-135 | 10 | 286 | 11 | 298 | |
| 6.91e-135 | 5 | 286 | 9 | 297 | |
| 1.02e-134 | 10 | 286 | 11 | 298 | |
| 1.90e-130 | 10 | 286 | 11 | 299 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.56e-120 | 27 | 232 | 1 | 206 | Acetylxylan Esterase From P. Purpurogenum Refined At 1.10 Angstroms [Talaromyces purpureogenus] |
|
| 6.32e-120 | 28 | 232 | 2 | 206 | ACETYLXYLAN ESTERASE AT 0.90 ANGSTROM RESOLUTION [Talaromyces purpureogenus] |
|
| 4.23e-118 | 27 | 232 | 1 | 206 | Iodinated Complex Of Acetyl Xylan Esterase At 1.80 Angstroms [Talaromyces purpureogenus] |
|
| 2.95e-111 | 28 | 233 | 2 | 207 | Chain A, ACETYL XYLAN ESTERASE [Trichoderma reesei],1QOZ_B Chain B, ACETYL XYLAN ESTERASE [Trichoderma reesei] |
|
| 2.95e-07 | 254 | 286 | 4 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.44e-128 | 10 | 286 | 11 | 302 | Acetylxylan esterase OS=Hypocrea jecorina OX=51453 GN=axe1 PE=1 SV=1 |
|
| 1.52e-126 | 8 | 286 | 6 | 295 | Cutinase 11 OS=Verticillium dahliae OX=27337 GN=VD0003_g7577 PE=1 SV=1 |
|
| 4.39e-121 | 3 | 232 | 5 | 233 | Acetylxylan esterase 2 OS=Talaromyces purpureogenus OX=1266744 GN=axe-2 PE=1 SV=1 |
|
| 2.41e-10 | 254 | 286 | 339 | 371 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
|
| 1.90e-09 | 254 | 286 | 336 | 368 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
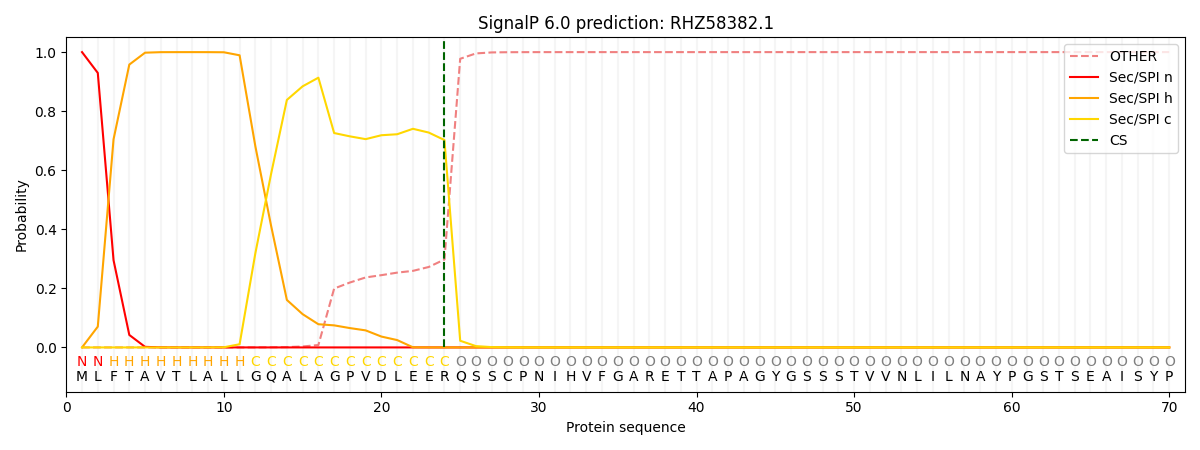
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000244 | 0.999724 | CS pos: 24-25. Pr: 0.7023 |
