You are browsing environment: FUNGIDB
CAZyme Information: RHZ54237.1
You are here: Home > Sequence: RHZ54237.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus thermomutatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus thermomutatus | |||||||||||
| CAZyme ID | RHZ54237.1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.15:80 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 53 | 378 | 1.1e-73 | 0.963076923076923 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395231 | Glyco_hydro_28 | 3.37e-129 | 58 | 379 | 1 | 321 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| 178580 | PLN03003 | 4.61e-29 | 117 | 328 | 120 | 326 | Probable polygalacturonase At3g15720 |
| 227721 | Pgu1 | 6.07e-27 | 94 | 304 | 183 | 408 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| 177865 | PLN02218 | 2.42e-21 | 55 | 372 | 92 | 417 | polygalacturonase ADPG |
| 215540 | PLN03010 | 8.19e-20 | 98 | 378 | 124 | 394 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.25e-224 | 4 | 379 | 5 | 382 | |
| 9.82e-221 | 18 | 379 | 18 | 379 | |
| 2.49e-218 | 1 | 379 | 1 | 378 | |
| 2.49e-218 | 1 | 379 | 1 | 378 | |
| 1.28e-213 | 6 | 379 | 7 | 385 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.62e-156 | 37 | 379 | 1 | 339 | Polygalacturonase From Aspergillus Aculeatus [Aspergillus aculeatus],1IB4_A Crystal Structure of Polygalacturonase from Aspergillus Aculeatus at Ph4.5 [Aspergillus aculeatus],1IB4_B Crystal Structure of Polygalacturonase from Aspergillus Aculeatus at Ph4.5 [Aspergillus aculeatus] |
|
| 8.84e-148 | 31 | 379 | 2 | 344 | Chain A, Endo-polygalacturonase [Evansstolkia leycettana] |
|
| 5.08e-147 | 31 | 379 | 2 | 344 | Chain A, endo-polygalacturonase [Evansstolkia leycettana] |
|
| 4.43e-146 | 40 | 379 | 3 | 336 | Chain A, Endo-polygalacturonase [Evansstolkia leycettana] |
|
| 1.99e-145 | 40 | 375 | 3 | 333 | Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini],2IQ7_B Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini],2IQ7_C Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini],2IQ7_D Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini],2IQ7_E Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini],2IQ7_F Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini],2IQ7_G Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins [Colletotrichum lupini] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.66e-222 | 1 | 379 | 1 | 378 | Probable endopolygalacturonase AFUB_016610 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_016610 PE=3 SV=1 |
|
| 5.66e-222 | 1 | 379 | 1 | 378 | Probable endopolygalacturonase AFUA_1G17220 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_1G17220 PE=3 SV=1 |
|
| 4.42e-219 | 1 | 379 | 1 | 378 | Probable endopolygalacturonase NFIA_008150 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_008150 PE=3 SV=1 |
|
| 4.14e-206 | 5 | 379 | 5 | 378 | Endopolygalacturonase AN8327 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=AN8327 PE=1 SV=1 |
|
| 4.06e-168 | 14 | 379 | 14 | 378 | Probable endopolygalacturonase E OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pgaE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
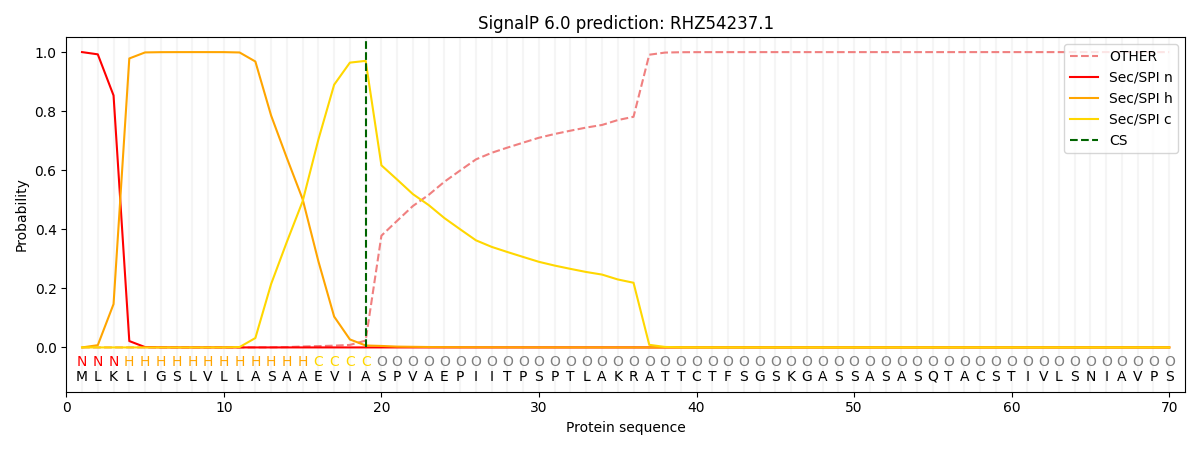
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000265 | 0.999708 | CS pos: 19-20. Pr: 0.9703 |
