You are browsing environment: FUNGIDB
CAZyme Information: RAK81986.1
Basic Information
help
| Species |
Aspergillus fijiensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis
|
| CAZyme ID |
RAK81986.1
|
| CAZy Family |
GT39 |
| CAZyme Description |
glycoside hydrolase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 334 |
KZ824623|CGC4 |
37038.88 |
3.8393 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AfijiensisCBS313.89 |
12336 |
1448319 |
318 |
12018
|
|
| Gene Location |
No EC number prediction in RAK81986.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
134 |
279 |
5.6e-20 |
0.43564356435643564 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 226609
|
ManB2 |
5.21e-06 |
144 |
263 |
177 |
298 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| 396639
|
Glyco_hydro_26 |
5.72e-04 |
191 |
277 |
197 |
276 |
Glycosyl hydrolase family 26. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.91e-58 |
39 |
308 |
34 |
291 |
| 4.48e-57 |
39 |
308 |
81 |
338 |
| 4.72e-53 |
18 |
334 |
14 |
330 |
| 3.01e-52 |
39 |
333 |
34 |
317 |
| 2.00e-50 |
57 |
315 |
48 |
286 |
RAK81986.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5.28e-07 |
123 |
261 |
167 |
297 |
Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
This protein is predicted as SP
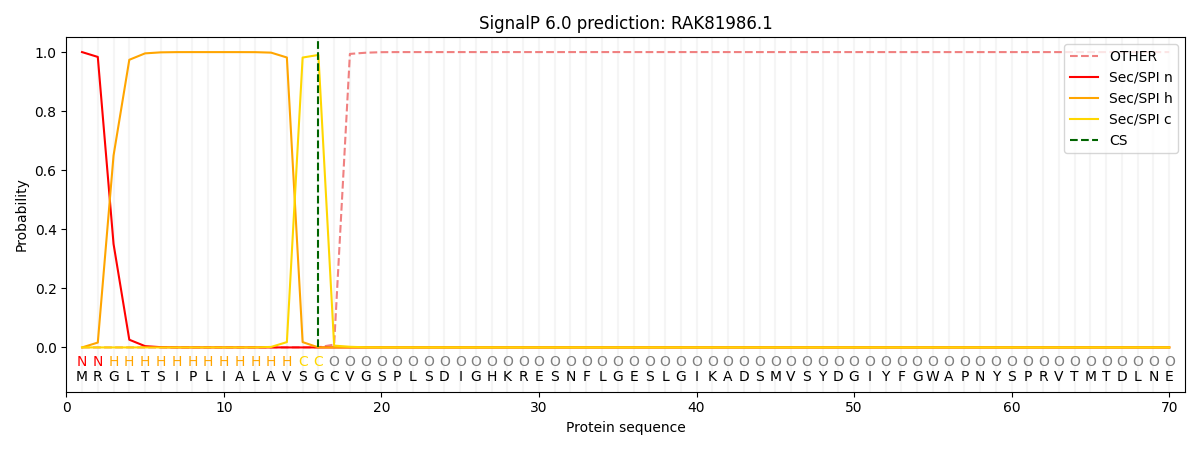
| Other |
SP_Sec_SPI |
CS Position |
| 0.000000 |
1.000053 |
CS pos: 16-17. Pr: 0.9903 |
There is no transmembrane helices in RAK81986.1.

